- Research
- Open access
- Published:
Tissue-specific usage of transposable element-derived promoters in mouse development
Genome Biology volume 21, Article number: 255 (2020)
Abstract
Background
Transposable elements (TEs) are a significant component of eukaryotic genomes and play essential roles in genome evolution. Mounting evidence indicates that TEs are highly transcribed in early embryo development and contribute to distinct biological functions and tissue morphology.
Results
We examine the epigenetic dynamics of mouse TEs during the development of five tissues: intestine, liver, lung, stomach, and kidney. We found that TEs are associated with over 20% of open chromatin regions during development. Close to half of these accessible TEs are only activated in a single tissue and a specific developmental stage. Most accessible TEs are rodent-specific. Across these five tissues, 453 accessible TEs are found to create the transcription start sites of downstream genes in mouse, including 117 protein-coding genes and 144 lincRNA genes, 93.7% of which are mouse-specific. Species-specific TE-derived transcription start sites are found to drive the expression of tissue-specific genes and change their tissue-specific expression patterns during evolution.
Conclusion
Our results suggest that TE insertions increase the regulatory potential of the genome, and some TEs have been domesticated to become a crucial component of gene and regulate tissue-specific expression during mouse tissue development.
Background
In the mammalian genome, only about 2% of DNA can be translated into protein products; the remaining ~ 98% of non-coding genome is considered to be “genomic dark matter” with unknown function [1,2,3]. Within these non-coding sequences, approximately 37% of the mouse genome and 45% of the human genome are derived from different kinds of transposable elements (TEs), including LINE, SINE, ERV, and DNA transposons [1,2,3,4,5,6]. TEs are highly repetitive DNA units and can reproduce themselves in the host genome. TEs generally belong to two main categories: DNA transposons, which mobilize themselves in the genome through a “cut and paste” mechanism; and retrotransposons, which replicate themselves by a “copy and paste” mechanism and can reach to hundreds of thousand copies in the genome [6, 7].
As parasites in the mammalian genome, TEs can regulate their activities by hijacking the regulatory mechanism of the host genome. Additionally, because of the highly abundant transcriptional factor binding sites in TE sequences, TEs also present a substantial regulatory potential to the host genome [8,9,10,11]. In order to resist the damaging effect of TEs, the host genome has evolved multiple mechanisms, in particular epigenetic repression, including DNA methylation and repressive histone methylation, to suppress the activity of TEs in the cell [12]. In the mammalian genome, most TEs are believed to be epigenetically silenced in somatic cells. However, mounting evidence suggests that some TEs escape from epigenetic silencing and are actively involved in a diverse array of biological processes (e.g., embryogenesis and carcinogenesis) and can become an essential component (e.g., promoter, enhancer, or insulator) of gene regulatory networks in the host genome [5, 7,8,9,10, 13,14,15,16,17]. Specifically, TEs have been found to initiate some genes’ transcription, especially for genes involved in immunity or response to external stimuli [18,19,20,21,22,23]. Recent evidence also suggests that TEs are significant contributors to the origin of vertebrate long non-coding RNAs [7, 24]. Meanwhile, TEs were also found to play roles as promoters in early development [25,26,27,28,29] and some terminally differentiated tissues [30, 31].
Activation of silent TEs is tightly associated with epigenetic modification [14, 32,33,34,35]. In our previous study [14], we reported the tissue-specific pattern of DNA methylation of 928 TE subfamilies across different human tissues. When TEs were unmethylated in a specific tissue, active enhancer histone modification marks, such as H3K4me1 and H3K27ac, were also enriched around these unmethylated TEs. We further found that these tissue-specific activated TEs can be associated with tissue-specific transcription factors (TFs), suggesting that the TFs might directly control the activation of TEs in a tissue-specific manner. We previously utilized the tissue-specific epigenetic landscape to detect the activation of TEs in human cancer cells [13, 14, 17, 35, 36]. In particular, we reported highly active cryptic transcription of LTR12 subfamilies when the lung cancer cell line NCI-H1299 underwent treatment of a DNMT inhibitor and an HDAC inhibitor [35]. Other studies also indicated the active roles of TEs in neurodegeneration [37] and systemic lupus erythematosus [38]. Altogether, such evidence suggests a critical need to better understand the function of TEs in different biological processes.
In this study, we analyzed the dynamics of chromatin accessibility in five mouse tissues [39, 40], including intestine, liver, lung, kidney, and stomach, to investigate the involvement and contribution of TEs to gene regulatory networks in mouse embryo development. We identified 73,453 accessible TEs in these five tissues, and ~ 53% of these TEs exhibited dynamic accessibility during tissue development. The activation of TEs was strongly associated with the developmental stage-specific transcription factors. The accessibility of TEs displayed a highly tissue-specific pattern, and genes around accessible TEs were associated with tissue-specific functions. During the development of five mouse tissues, 453 TEs were found to create the new 5′ end TSS of genes, including 117 protein-coding genes and 144 lincRNA genes. 13.8% of these genes with TE-derived TSSs showed a tissue-specific expression pattern. Thus, mouse-specific TEs have been domesticated and provided functional promoters for downstream protein-coding genes and have created a novel tissue-specific expression pattern for these genes. Taken together, our study provides a comprehensive investigation into the contribution of TEs to the regulatory landscape of mouse embryonic tissue development.
Results
The tissue-specific pattern of accessible TEs in mouse tissue development
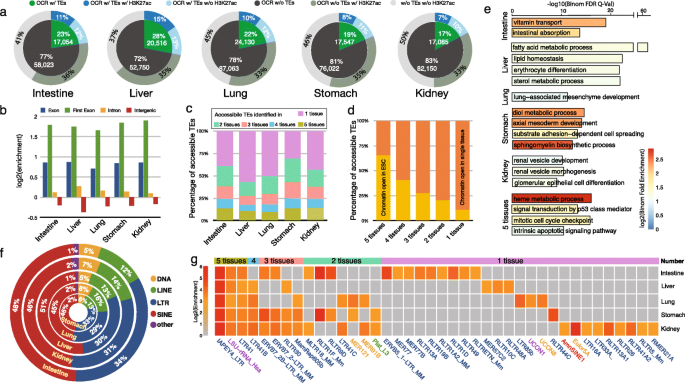
To understand the functionality of TEs during mouse embryonic development, we obtained the epigenomic data of five tissues, including intestine, liver, lung, kidney, and stomach, at two developmental stages: embryonic day 14.5 (E14.5) and postnatal day 0 (P0) from the ENCODE [41, 42] data portal (Additional file 1). By analyzing ATAC-seq data from these five mouse tissues, we identified 452,298 open chromatin regions (OCR) in total and examined the accessibility of TEs during tissue development and found ~ 21% of these open chromatin regions were associated with TEs (Additional file 2). Although spatial correlation analysis calculated by GenometriCorr [7, 43] indicated the significant lack of overlap between OCRs and TEs compared with random expectation (Additional file 3), consistent with the general notion that TEs are epigenetically silenced, the sheer number of accessible TEs underscoresd their potential sizable contribution to gene regulation [34, 44, 45]. In each tissue, TEs were associated with 17% (kidney) to 28% (liver) of OCRs, and about half of these accessible TEs directly overlapped with the active histone modification H3K27ac (Fig. 1a; Additional file 4: Table S1). Most accessible TEs (> 90%) were located in intragenic or intergenic regions (Additional file 4: Table S2; Additional file 5: Fig. S1), suggesting that these accessible TEs could be potential enhancers playing a role in tissue development. Interestingly, we noticed that ~ 6% of accessible TEs overlapped with exons and were especially enriched in the first exons comparing with the random background (Fig. 1b). Such results suggested the potential role of TEs as promoters in mouse tissue development as previously reported [7, 19,20,21,22,23,24, 46,47,48,49,50,51]. Approximately 10% of accessible TEs were commonly accessible in all five tissues (Fig. 1c), and they were enriched for CTCF, Usf1, and Klf4 binding motifs (Additional file 4: Table S3 and S4). Close to half of accessible TEs were only accessible in a single tissue type (Fig. 1c; Additional file 4: Table S3). These results confirmed previous reports suggesting that the TE activation displayed a strong tissue-specific pattern [14, 24, 28, 52,53,54].
Accessible transposable elements (TEs) in five mouse tissues. a Around 17~28% of ATAC-seq peaks identified in five mouse tissues associated with TEs, near half of these accessible TEs recruited H3K27ac signals. b Genomic distribution enrichment of accessible TEs of five mouse tissues in the mouse genome. Accessible TEs highly enriched in the exons of the mouse genome, especially the first exon. c Distribution of accessible TEs among five mouse tissues. Near half of accessible TEs were identified in only one tissue type. d Chromatin status of identified accessible TEs in mouse ES cells. In total, 65% of accessible TEs constitutively in five tissues were accessible in mouse ESC, only 10% of tissue-specific TEs were open in mESC. e Gene ontology (biological process) enrichment analysis of tissue-specific accessible TEs in five mouse tissues. GO terms related to tissue-specific function were selected to visualize. f TE class distribution of accessible TEs in five mouse tissues. g TE subfamily enrichment of accessible TEs in five mouse tissues
Next, we asked whether these accessible TEs in developing tissues were also accessible in the mouse embryonic stem cells (mESCs). Interestingly, the majority of the constitutively accessible TEs (65%) already had open chromatin accessibility in mESCs, whereas only 10% of the tissue-specific accessible TEs had open chromatin accessibility in mESCs (Fig. 1d; Additional file 4: Table S5). Thus, the small set of TEs with early and constitutive open chromatin signatures might play a more general regulatory role in development, whereas TEs exhibiting late and tissue-specific open chromatin might be related to tissue-specific gene regulation. Consistent with this hypothesis, our GO enrichment analysis revealed distinct functional enrichment patterns among genes associated with different classes of accessible TEs (Fig. 1e; Additional file 6). The constitutively accessible TEs were more associated with essential biological processes and functions, such as ribosome biogenesis and mitotic cell cycle checkpoint, whereas tissue-specific accessible TEs were more associated with functions directly relevant to the specific tissue types (Fig. 1e; Additional file 6). Close to half of the accessible TEs belonged to the SINE class, and ~ 30% were derived from LTR elements. Of the remaining, 15% were LINEs, and less than 8% of accessible TEs belonged to DNA class (Fig. 1f; Additional file 4: Table S6). However, subfamily enrichment analysis of accessible TEs indicated that most of the top enriched TE subfamilies were LTRs (Fig. 1g; Additional file 4: Table S7). Three TE subfamilies (IAPEY4_LTR, LSU-rRNA_Hsa, and LTR41) were highly enriched in constitutively accessible TEs (Fig. 1g; Additional file 4: Table S7). LTR41b, an LTR subfamily exhibiting strong regulatory ability in human liver cancer cell line HepG2 [55], was highly accessible in four mouse tissues but not the intestine.
TEs can initiate the transcription of genes during tissue development
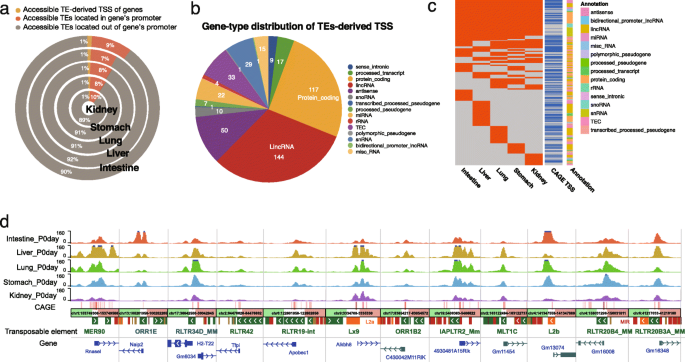
Within the 6% of accessible TEs that overlapped exons, 60% of them overlapped with the first exon of annotated genes (intestine 59.9%; liver 57.5%; lung 60.6%; stomach 62.7%; kidney 64.2%). This result motivated us to examine the relationship between these TEs and the promoter or the transcription start site (TSS) of the overlapping gene. We found ~ 10% of accessible TEs were located in the promoter regions (0.5 kb upstream of the TSS) of genes (Fig. 2a; Additional file 7: Table S1). In particular, we found that about 1% of accessible TEs directly overlapped with the transcription start site of genes, suggesting that TEs might have been domesticated as an integral component of the downstream gene for RNA Polymerase II recruitment and transcription initiation (Fig. 2a). Across the five developing mouse tissues, 453 TEs were found to derive the TSS of genes, including 117 protein-coding genes, 144 lincRNA genes, and 50 antisense genes (Fig. 2b; Additional file 7: Table S2). In total, 58% of these accessible TEs can be independently validated by the FANTOM5 CAGE-TSS signal. A total of 46% of the TEs that can derive the TSS of a gene was only accessible in one tissue, only slightly smaller than the overall tissue specificity of accessible TEs (Fig. 2c; Additional file 7: Table S2). In contrast, ~ 20% of TEs-derived TSSs were constitutively accessible, much higher than the 10% overall constitutively accessible TE percentage (Fig. 2c, d; Additional file 7: Table S2 and S3; https://bit.ly/3hZryCj).
Genes with accessible transposable elements (TEs)-derived TSS in five mouse tissues. a Distribution of accessible TEs in the gene’s promoter and TEs-derived TSS in five tissues. Near 10% of accessible TEs located in promoter regions, and 1% accessible TEs contained gene TSS. b Gene-type distribution of 453 accessible TE-derived TSSs identified in five mouse tissues, including 117 protein-coding genes and 144 lincRNA. c Tissue-specific distribution of 453 accessible TE-derived TSSs. A total of 263 of these accessible TEs can be validated by CAGE-seq. The annotation colors represented different gene types. Near half of TE-derived TSSs were only identified in one tissue. d Epigenome browser view of ATAC-seq signals around the accessible TE-derived TSS of protein-coding genes (left six) and lincRNA (right six)
Dynamically accessible TEs were associated with developmental stage-specific transcription factors
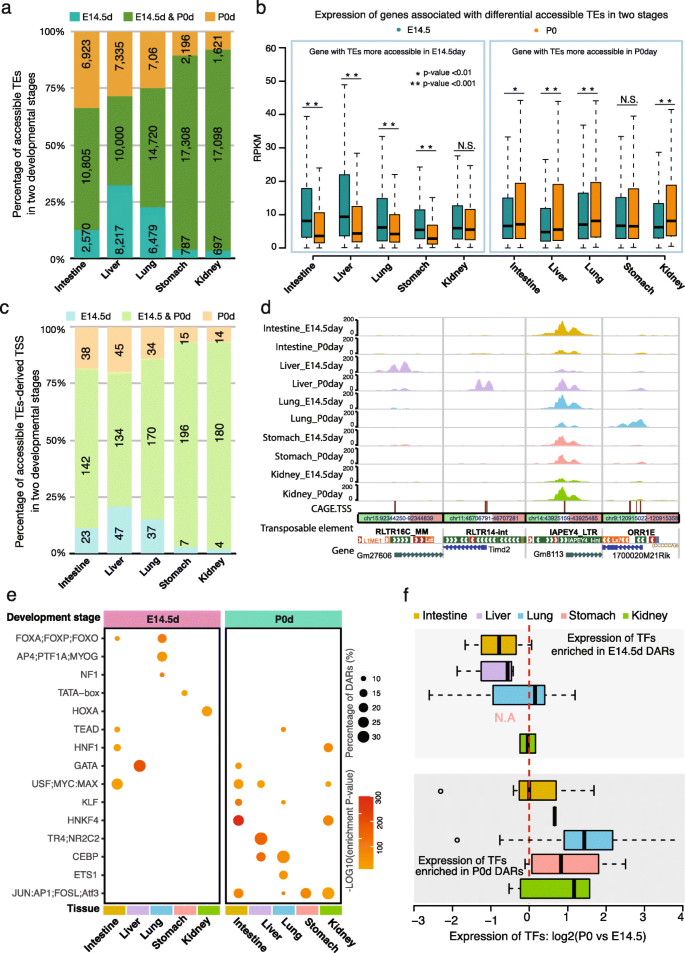
Since we incorporated two developmental stages (embryo day 14.5 and postnatal day 0) of five tissues in our analysis, we were able to explore the dynamic accessibility changes of TEs during mouse development. In total, we found that the accessibilities of 38,560 TEs were developmental stage-dependent. In the intestine, liver, and lung, about 50% of accessible TEs showed changes of accessibility between the two developmental stages (Fig. 3a; Additional file 8: Table S1). In contrast, only about 10% of accessible TEs showed changes in the stomach and kidney between E14.5 and P0, suggesting that stage-specific TE accessibilities were also tissue-specific (Fig. 3a). Genes associated with developmental stage-specific accessible TEs generally also displayed developmental stage-specific expression pattern (Fig. 3b). Since ~ 95% of developmental stage-specific accessible TEs were distal to gene’s promoter, our results supported previous findings that TEs might play roles as a distal enhancer in gene regulatory networks (Additional file 8: Table S2) [9, 34, 48, 56]. Furthermore, 214 TE-derived TSSs of genes exhibited accessibility changes (Fig. 3c; Additional file 8: Table S1). In the liver, 47 TE-derived TSSs were accessible only at E14.5, and 45 TE-derived TSSs were accessible at P0. In contrast, the numbers for the kidney were 4 and 14 for E14.5 and P0, respectively (Fig. 3c). Just to highlight a few examples, Gm27606 (RLTR16C derived) and Timd2 (RLTR14-int) were only accessible in the liver, but at different developmental stages; 1700020M21Rik (ORR1E) only became accessible in the lung at P0; Gm8113 (IAPEY4_LTR) was accessible across all five tissues but in a developmentally dependent manner: early in intestine and lung, but late in liver and kidney (Fig. 3d; https://bit.ly/2EVYLjX).
Dynamic accessibility changes of TEs during mouse embryonic development. a Percentage of dynamically changed accessible TEs at two development stages in five mouse tissues. E14.5d: the accessible TEs with higher accessibility in the E14.5 development stage; P0d: accessible TEs with higher accessibility in P0day; E14.5&P0d mean accessible TEs did not significantly change their accessibility between the two stages. b Expression pattern of genes around accessible TEs at two development stages in five mouse tissues (Wilcoxon signed-rank test). c Percentage of dynamically changed accessible TE-derived TSS at two development stages in five mouse tissues. d Epigenome browser view of ATAC-seq signals around the dynamically changed accessible TE-derived TSS of protein-coding genes (left three) and lincRNA (right one). e Tissue- and developmental stage-specific enrichment of predicted transcription factors binding motifs in dynamically changed accessible TEs at E14.5 (left) and P0 (right) in five mouse tissues. The color of the dot indicated the enrichment p-value of motif; the size represented the percentage of DARs that contained the TF motif. f Developmental stage-specific expression pattern of TFs that predicted to bind the dynamically changed accessible TEs at E14.5 (top) and P0 (bottom) in five mouse tissues. The dashed line indicated no expression difference of TFs between two stages. N.A: no TF motif was enriched in stomach P0d DARs
Our previous study revealed a tight connection between activation of TEs and the binding of transcription factors [14, 57]. Based on motif enrichment analysis in the developmental stage-specific accessible TEs, we found that the TF binding motifs (TFBS) were stage-specifically enriched (Fig. 3e; Additional file 8: Table S3 to S7). To understand the expression pattern of these TFs in two distinct developmental stages, we further analyzed the corresponding RNA-seq data and examined the expression level of these TFs in all five tissues. As expected, we noticed a developmental stage-specific expression pattern of enriched TFs (Fig. 3f). For example, we found that the GATA binding motif was enriched in both intestine and liver at different developmental stages. Previous studies revealed the importance of GATA4 and GATA6 in the development of both intestine and liver [58,59,60,61,62,63]. In our study, we observed that GATA5 and GATA6, but not GATA4, were upregulated in intestine development (Additional file 8: Table S8). Moreover, we found GATA1 had the highest expression in the GATA family (~ 20 fold higher than GATA4 and GATA6) in the liver at E14.5 and was most significantly downregulated at P0 stage (Additional file 8: Table S8). This result suggested that GATA1 might also play important roles in regulating accessible TEs and contribute to liver development. We also found the motif of bZIP TFs, including Jun, Junb, Batf, and Atf3, was highly enriched in multiple tissues at P0. Correspondingly, the expression of these TFs was upregulated during embryo development (Additional file 8: Table S8).
Accessible TEs altered the tissue specificity of gene expression
Next, we examined the evolutionary conservation of accessible TEs identified in our study. Based on current genome annotation and alignment [64], we found 10% of mouse TEs had orthologous counterparts in the human genome, and 65% had orthologous sequences in the rat genome. For accessible TEs, these two numbers were 30% and ~ 87% (Fig. 4a; Additional file 9: Table S1). In contrast, close to 75% of the non-TE open chromatin regions in the mouse genome had orthologs in the human genome, and 95% had orthologous sequences in the rat genome (Fig. 4a; Additional file 9: Table S1). These results supported a previous finding that regulatory elements, including TE-derived elements, were more likely to be retained in the genome [65]. By comparing the 60-ways PhastCons conservation of accessible TEs with that of their surrounding genomic sequence, we found that accessible TEs had much lower PhastCons scores than gene-coding exon, OCR without TEs, and intronic regions (Fig. 4b). These results suggested that most of the mouse accessible TEs were putative rodent-specific regulatory elements and were much less conserved compared to non-TE regulatory elements, at least based on sequence alignment-based estimates.
Evolutionary conservation of mouse accessible TEs in the rat and human genome. a The distinct proportion of orthologous mouse accessible TEs in the rat and human genome (left), orthologous mouse background regions in the rat and human genome (middle), and orthologous mouse OCRs without TEs in the rat and human genome (right). b Averaged phastCons score at 10-kb regions centered by accessible TEs (red), OCRs without TEs (blue), gene exon (orange), and intron (gray) in the mouse genome. Accessible TEs showed the lowest phastCons score in the center. c Distinct orthologous conservation of protein-coding genes (left) and lincRNA (right) with TE-derived TSS. In total, 87% of mouse protein-coding genes with TE-derived TSS were ortholog in both human and rat genome, while less than 1% of mouse lincRNAs were ortholog in other two species. d Expression pattern of 117 protein-coding genes with mouse TE-derived TSS in five tissues of mouse and human. e Epigenome browser view of ATAC-seq signals and genome-alignment (mm10, hg38, and rn5) around rodent-specific ORR1A4 TE-derived TSS (left: conserved in mouse and rat) and canonical ortholog TSS (right: conserved in mouse, rat, and human) of Chit1 gene. Top-left: expression pattern of Chit1 in five mouse tissues. Top-right: expression pattern of CHIT1 in five human tissues. ANOVA test: p-value < 0.01
We further examined the conservation of accessible TEs that contained the 5′ end TSSs of genes (Fig. 4c; Additional file 9: Table S2). Consistent with overall accessible TEs, these TSS-deriving TEs were also mostly rodent-specific (59%). However, their downstream genes exhibited a more complex evolutionary pattern. Protein-coding genes were highly conserved: 81% of the 117 protein-coding genes with TE-derived TSSs had orthologs in all three species; only 18 did not have orthologs in the human genome, and only four were mouse-specific (Additional file 9: Table S3). In stark contrast, 94% of the 114 lincRNA genes with TE-derived TSSs were mouse-specific (Additional file 9: Table S4). This result was consistent with previous studies that suggested that species-specific TEs contributed to the origination of lincRNAs during evolution [7, 50, 66, 67].
Next, we asked how these domesticated TSS-deriving TEs influenced expression of their host genes. We compared the expression pattern of 95 conserved protein-coding genes across five tissues of mouse and human. We found more than half of these protein-coding genes showed distinct expression patterns at the tissue level between human and mouse (Fig. 4d). This result suggested that the domestication of TEs as a TSS may provide a mechanism to allow new tissue-specific gene expression to evolve. For example, Chit1 encodes an enzyme called Chitinase 1, which is secreted by activated macrophages and plays a role in the degradation of chitin-containing pathogens [68, 69]. In the mouse genome, the 5′ TSS of Chit1 is located in ORR1A4, which is a rodent-specific TE and not present in the human genome (Fig. 4e). This TE had strong accessibility in the mouse liver and stomach, but relative weak accessibility in the lung. Transcriptome analysis indicated Chit1 was expressed the highest in mouse stomach (Fig. 4e; ANOVA test: p-value < 0.01; Additional file 5: Fig. S2). Interestingly, we did not detect Chit1 expression in the RNA-seq data of mouse liver, possibly because the liver still missed certain cooperating transcription factors to initiate the transcription. Then, 27 kb downstream from this TE-derived TSS lied the canonical TSS, which is orthologous to the TSS of the human CHIT1 gene. Interestingly, we did not observe any ATAC-seq signal around this conserved ortholog TSS in any of the five mouse tissues in our study (Fig. 4e), suggesting that this alternative TSS was not used in mouse genome for transcription initiation. In the corresponding five human tissues, CHIT1 was only expressed in the lung, which contains a large population of alveolar macrophages [70]. We reasoned that the conserved TSS represented the ancestral element and was responsible for CHIT1 expression in human lung tissue. However, the integration of ORR1A4 upstream of the canonical TSS in the rodent genome created a stronger TSS and a new first exon to initiate the transcription of Chit1 in the stomach, potentially due to the sequence features of ORR1A4 that could respond to the stomach-specific transcription factors. Thus, a transposable element integration event not only resulted in the promoter turnover of nearby genes, but also initiated the gene transcription in a novel tissue.
Domesticated TEs created tissue-specific expression of species-specific genes
About 19% of protein-coding genes that utilized an accessible TE-derived TSS were rodent- or mouse-specific (Fig. 4c). We noticed that 11 of these 22 rodent- or mouse-specific protein-coding genes were expressed in a tissue-specific manner (Fig. 4d). Timd2 (T cell immunoglobulin and mucin domain-containing 2), a gene that can enhance T cell activation, was found to use an RLTR14-int element to initiate its liver-specific expression. Timd2 is a rodent-specific gene, which originated from a genome duplication event in the mouse and rat genomes. A 230-kb-long DNA fragment containing Timd2, Dppa1, and several non-coding genes were duplicated and positioned inversely in the mouse genome between Havcr2 and Havcr1 (Fig. 5a). Timd2 was a paralog of Havcr1 resulting from the genome duplication. Both Timd2 and Havcr1 encode a 305aa protein with 60% identity at the amino acid level and 74% identity at the coding DNA level (Fig. 5b; Additional file 5: Fig. S3). Havcr1 (Hepatitis A virus cellular receptor 1) is also called KIM-1(Kidney Injury Molecule 1), and is highly upregulated in injured kidneys [71]. HAVCR1 belongs to TIM family, which also includes HAVCR2 and TIMD4. As receptors for phosphatidylserine, TIM proteins bind many families of viruses such as hepatitis A, dengue, and ebola. We confirmed that Havcr1 was highly expressed in normal kidney across human, mouse, and rat. However, Timd2 was only expressed in mouse liver but not in the kidney (Fig. 5c; Additional file 5: Fig. S4; ANOVA test: p-value < 0.01).
Ortholog analysis of Timd2 gene. a Genome structure of Havcr2-Timd2-Havcr1 in the mouse genome (top) and genome structure of HAVCR2-HAVCR1 in the human genome (bottom). A 230-kb-long DNA fragment contained Timd2 gene only exists in the mouse genome. b Gene ortholog analysis of Havcr1, Timd2, and Timd4 in human, mouse, and rat based on protein sequence. Havcr1 genes from mouse and rat were firstly clustered with Timd2 genes instead of HAVCR1 gene in the human genome. c Expression pattern of Havcr1 and Timd2 in five tissues of human (left) and mouse (middle and right). ANOVA test: p value < 0.01. d Pair-wise alignment of exons between Havcr1 (top) and Timd2 (bottom). RLTR14-int-derived Exon1 of Timd2 did not have ortholog exon in Havcr1. e Top: Epigenome browser view of ATAC-seq and ChIP-seq signals around RLTR14-int-derived TSS of Timd2 in mouse liver. The signals of ATAC-seq, H3k27ac, and TFs binding were co-localized at Timd2 TSS region within RLTR14-int element. Bottom-left: expression pattern of Rxra in five mouse tissues. Rxra showed the highest expression in the liver. ANOVA test: p-value < 0.01. Bottom-right: predicted TF binding motifs within Timd2-TSS RLTR14-int element. f Pair-wise alignment among 128 RLTR14-int copies (overlapped to RLTR14-int-derived Timd2 TSS) against to RLTR14-int consensus sequence. Top: Gene structure of full-length consensus sequence of RLTR14-int (5317 bp). Middle: 661 bp pair-wise alignment between consensus sequence and Timd2-TSS RLTR14-int element. Six TF binding motifs were identified at the exactly same positions in both consensus sequence and Timd2-TSS RLTR14-int element. Bottom-left: 661 bp pair-wise alignment and conservation of consensus sequence (1st row), Timd2-TSS RLTR14-int element (2nd row), and 127 RLTR14-int copies. Bottom-right: occupancy of six TFBSs matched to pair-wise alignment
A close examination of the pair-wise alignment between mouse Timd2 and Havcr1 revealed the homology between the two genes as well as a Timd2-specific first exon, which was derived from a rodent-specific RLTR14-int element (Fig. 5d; Additional file 10). Transcription factor binding motifs, including Hnf4a, Cebp, Tfap2, and RxRa, were predicted to be present in this RLTR14-int element, and the element was accessible and displayed strong RNA Polymerase II binding (Fig. 5e). ChIP-seq data also confirmed the binding of transcription factors, including Hnf4a, Cebpb, and Rxra, to the RLTR14-int (Fig. 5e). Rxra exhibited a strong liver-specific expression pattern, whereas Hnf4a and Cebpb expressions were more ubiquitous across all the tissues (ANOVA test: p value < 0.01). These results suggested that following the genomic duplication that created Timd2, the integration of RLTR14-int created an alternative promoter of Timd2 that rewired its liver-specific expression pattern, likely by responding to liver-specific transcription factors such as Rxra (Fig. 5e).
Motif analysis of the RLTR14-int consensus sequence revealed six TF binding motifs (SCRT2/HLTF, STAT1::STAT2, NFIC::TLX1, TCF2, Sox3, Pparg::Rxra) in the region corresponding to new Timd2 promoter derived from the RLTR14-int element (Fig. 5f). All six TFBSs were well conserved in the RLTR14-int element that gave rise to Timd2 promoter. Alignment of 1451 RLTR14-int genomic fragments to the consensus sequence identified another 127 RLTR14-int copies that partially overlapped with the same region that derived Timd2 promoter. However, none of the 127 copies contained the full set of six TFBSs identified in the consensus sequence (Fig. 5f), although some of them were accessible TEs in different tissues (Additional file 5: Fig. S5). These results suggested that the key TFBSs were probably already present in the ancestral RLTR14-int, and the element that produced Timd2 promoter likely inherited the conserved TFBSs from the ancestral retrovirus.
Having identified a substantial contribution of TEs to evolving new TSSs, we next systematically determined the usage of the TE-derived TSS in comparison to the usage of alternative, non-TE TSS of the same 453 genes. Towards this end, we performed full-length transcript assembly and calculated each transcript’s expression level in five tissues at two different developmental stages. Of the 453 genes with TE-derived TSS, 316 genes had detectable expression in at least one tissue (> 0.1 RPKM). In total, 193 (61%) of these genes used 5′ end TE-derived TSS exclusively to initiate transcription, e.g., Cd302 and Ms4a4d (Additional file 5: Fig. S6A and S6B). A total of 107 (34%) of them utilized both 5′ end TE-derived TSS and non-TE TSS. Only 16 (5%) did not use 5′ end TE-derived TSS in these five tissues (Fig. 6a; Additional file 11: Table S1). We also assessed the contribution of 5′ end TE-derived TSS in producing transcripts and found that most genes had a higher fraction of their total transcripts originated from 5′ end TE-derived TSS across all five tissues and two developmental stages (Fig. 6b).
Contribution of 5′ end TE-derived TSS in tissue-specific gene expression. a Usage of 5′ end TE-derived TSS of 300 genes expressed in at least one tissue (> 0.1 FPKM). b Contribution of 5′ end TE-derived TSS in gene expression at two developmental stages in five mouse tissues. Y-axis: the expression percentage of 5′ end TE-derived transcripts contributed to the total gene’s expression at both E14.5 and P0 stage in each tissue. c Usage of 5′ end TE-derived TSS of 107 genes that used both 5′ end TE-derived and non-TE-derived TSS in five tissues at two developmental stages. The color of each dot represented the expression percentage of 5′ end TE-derived transcripts, and the size indicated the total gene expression values (FPKM). d Assembled transcripts of Tfpi2 genes at the P0 stage in five mouse tissues. Top: Sashimi plot visualization of splicing-junctions from RMER5-derived 1st exon to next exon in intestine, liver, and kidney. Bottom: expression of three assembled Tfpi2 transcripts across five tissues. e Assembled transcripts of Timd2 genes from E11.5 to P0 stages in mouse liver. Top: Sashimi-plot visualization of splicing-junctions from RLTR14-int-derived 1st exon to next exon in all seven developmental stages. Bottom: expression of RLTR14-int-derived Timd2 increased 13-fold across seven developmental stages in mouse liver
To further quantify the pattern of differential usage of 5′ end TE-derived promoter, we compared the expression level of transcription initiated from 5′ end TE-derived promoter with the non-TE-derived promoter for the 107 genes which utilized both types of promoters. A little more than half (55/107) of the genes used non-TE-derived promoters more often, and when they did so, the pattern was uniform across all five tissues. In contrast, the remaining half (52/107) used 5′ end TE-derived promoters more often, and when they did so, the pattern was tissue-specific (Fig. 6c). Tfpi2, a tumor suppressor gene and a member of Kunitz-type serine proteinase inhibitors [72], was found to use an RMER5-derived promoter to initiate 76% of gene transcription in the liver, but this promoter was not used in the lung and stomach at all (Fig. 6d). Car1, a carbonic anhydrase involving erythrocytes oxygen release, was specifically expressed in the developmental liver via a MER74C-derived promoter (Additional file 5: Fig. S6C). Finally, we observed that the usage of TE-derived promoters could be associated with developmental stages. Naip1, a NLR family gene encoding apoptosis inhibitor protein 1, was only transcribed in the P0day intestine by using an ORR1E-derived promoter (Additional file 5: Fig. S6D). The expression of Timd2 in the liver increased more than 13 times from E11.5day to P0day, and the RLTR14-int derived promoter contributed almost exclusively to this increase (Fig. 6e).
To provide an orthogonal validation of discovery we made from the transcript assembly of RNA-seq data, we analyzed the CAGE data of five matching tissues generated by FANTOM5 [73]. In total, 114 of 453 candidate genes expressed in five mouse tissues during embryo development, and 75 of them had CAGE peaks at TE-derived promoters (Additional file 11: Table S2). By assessing the expression level of TE-derived promoter and non-TE-derived promoter for each gene using CAGE data, we found that about half of the genes only used the TE-derived promoter to initiate gene expression at both E14.5 and P0 (Additional file 5: Fig. S7A, S7B). Of the 18 genes with multiple TSSs, eight were predominantly using their TE-derived promoters to initiate gene expression based on CAGE signals (Additional file 5: Fig. S7C). Importantly, we found very high concordance between CAGE-seq signal and assembled RNA-seq expression in TE-derived promoter usage (Additional file 5: Fig. S7D).
Discussion
Recent high-throughput sequencing technologies to characterize the transcriptomes and epigenomes of multicellular eukaryotes have revealed the crucial roles of TEs in the evolution of gene regulation. Integration of TEs into host genomes has brought numerous TF binding sites, and such perturbation of the genome interfered with gene regulatory networks and increased the regulatory dynamics of the genome when responding to external stimuli [18,19,20,21,22,23]. Specifically, the intact transcriptional elements of TEs can create novel transcription start sites (TSSs) to initiate transcription in the host genome. These novel TSSs were found to be the origination of a large proportion long non-coding RNAs [24, 46, 48, 50, 51, 66] and also created novel transcript isoforms of conserved protein-coding genes during evolution [8, 17, 20, 37, 74, 75].
Here we conducted a study to understand the functionality of TEs during mouse embryo development and focused on the activation of TE-derived TSSs in the development of five tissues. Leveraging the public datasets generated by ENCODE, we analyzed the epigenome data of five tissues (intestine, liver, lung, stomach, and kidney) at two different developmental stages: embryonic day 14.5 (E14.5) and postnatal day 0 (P0) (Additional file 1). On average, we observed that about 21% of the OCR in mouse development could be derived from TEs, suggesting TEs could escape from epigenetic silencing of the host genome during evolution. We noticed that liver had more accessible TEs, which account for 28% of the regulatory regions during development, when compared to the other four tissues (Fig. 1a). Such a phenomenon might be associated with the distinct evolutionary pressure and speed of gene regulatory network evolution in different tissues [76,77,78].
Regulatory elements, especially enhancers, are activated in a tissue-specific pattern [13,14,15, 28, 34, 79, 80]. In our study, we observed that accessible TEs showed highly tissue-specific patterns. Only ~ 10% of total accessible TEs were found to be accessible in all five tissues (Fig. 1c), and most of these constitutively accessible TEs were already accessible in mouse embryonic stem cells (Fig. 1d). Such results suggested that these TEs might be involved in very early-stage development and associated with fundamental biological processes, such as cell cycle regulation, and support findings of previous studies [14, 24, 28, 32, 33, 52, 53]. To explore the roles of TEs in tissue development, we examined the differential accessibility of TEs by comparing E14.5 and P0 development stages for each tissue. Interestingly, most accessible TEs in the stomach and kidney did not show differential accessibility between these stages; this might reflect different developmental timing between the mouse stomach, kidney, and other tissues. Conversely, in the intestine, lung, and liver, nearly half of accessible TEs showed distinct chromatin accessibility between the two development stages (Fig. 3a) and reflected a significant change of gene regulation at a later embryonic developmental stage. Previous studies indicate that transcription factors (TFs) play important roles in tissue development and are highly enriched in the open chromatin regions [16, 34, 56, 67, 81,82,83]. In our study, we identified 15 enriched TFBSs in the developmental stage-specific accessible TEs. We further observed the developmental-specific expression pattern of these TFs in the five tissues. Similar to previous studies [9, 34, 48, 52, 56, 82], our results indicated the importance of TEs in gene regulatory networks, especially in the formation of tissue specificity. Such results suggested that the accessible TEs were regulated by developmental stage-specific TFs, including FOXA and NF1, which have been reported to play essential roles in the development of the lung [84,85,86,87].
In addition to distal accessible elements that are far from gene promoters, we found about 10% of accessible TEs can serve as the transcription start site (TSS) of a gene. Our study identified 453 accessible TE-derived 5′ end TSSs across the five tissues, and most of these TE-derived TSSs were validated by independent CAGE-seq data generated by the FANTOM5 project [73, 88]. Previous studies reported that TEs were major contributors to the origin of non-coding RNAs [24, 46,47,48,49,50,51]. In addition to confirming TE’s contribution to lincRNA, our results revealed a crucial evolutionary mechanism by which TEs contributed significantly to evolving alternative transcription start sites for protein-coding genes and, in some cases, to evolving novel tissue-specific expression patterns for these genes. We found that 95 of 117 protein-coding genes with TE-derived TSSs had orthologous genes in the rat and human genome.
Interestingly, we found that most of these accessible TSS TEs were rodent or mouse-specific: 66 of the 95 conserved protein-coding genes used a rodent/mouse-specific TE-derived TSS to initiate transcription in different tissues (Additional file 9: Table S3). This result highlights that regulatory elements can evolve much more rapidly than genes themselves. By evolving a new TSS or a new promoter, a gene can rapidly explore a variety of expression patterns across cell types, and potentially offer phenotypic diversity upon which selection can act. Here, transposable elements provide convenient building blocks for the evolution of new promoters, and thus presenting a unique opportunity to accelerate regulatory evolution for the species that they invade. For example, a rodent-specific ORR1A4 initiates stomach-specific transcription of the Chit1 gene. Chit1 is secreted by activated macrophages and plays a role in the degradation of chitin-containing pathogens [68, 69]. The active expression of Chit1 in the stomach might help the newborn mice to defend against pathogens encountered during feeding in early life. However, whether the expression of Chit1 comes from macrophages or stomach cells, such as parietal cells or mucous neck cells, still needs further investigation via other experimental approaches, such as single-cell RNA sequencing analysis or in situ hybridization.
We also found that TEs can initiate the novel expression pattern of rodent-specific genes. Timd2 is a rodent-specific gene that could enhance T cell activation by interacting with SEMA4A. Timd2 is the duplicated gene of Havcr1, which is highly conserved and expressed in the kidney across mammals, including mouse, rat, and human. Conversely, the Timd2 gene is only identified in the rodent genome and is especially highly expressed in the liver. Our analysis indicated that the insertion of the RLTR14-int element created a new 5′-end exon and initiated the transcription of Timd2 in mouse liver by recruiting liver-specific TFs, which can be inferred by motif analysis and ChIP-seq assays (Fig. 5e). Another interesting finding is that the Timd2 did not use the ancestral TSS that mainly initiates the expression of the Havcr1 gene in the kidney. We noticed that two SINE elements, B1_Mm and B1_Mus2, inserted around the ancestral TSS of Timd2. Our motif analysis revealed that several repressive TF binding sites, including those of SNAI2, FOXP1, and FOXD3, were present in these two SINE elements (Additional file 5: Fig. S8). Previous studies indicated KRAB-ZFPs could repress the activity of LTRs through histone modification and DNA methylation, and further repressed the expression of nearby genes in both ES cells and adult tissues [89, 90]. Thus, it is tempting to hypothesize that the integration of the two SINE elements around the ancestral TSS of Timd2 resulted in the silencing of that TSS, which indirectly facilitated the RLTR14-int-derived TSS to take over the expression of Timd2.
Conclusions
TEs are significant contributors to genome evolution, and the percentage of TEs in the genome is positively correlated with the size of the genome [91]. Species-specific TEs are found to play roles as regulatory elements in human and mouse [5, 7, 18, 20, 34, 92]. Our study not only confirms previous findings, but also extends the contribution of TEs to the initiation of gene transcription, especially to creating novel tissue-specific gene expression patterns by acting as alternative TSSs. We additionally found that TEs may also silence genes through the repressive mechanisms of the host genome. Further investigation is required to confirm such a hypothesis. In summary, our study provides a comprehensive investigation of TEs’ function during mouse development. We report that TE-derived alternative TSSs of protein-coding genes can drive distinct tissue-specific expression patterns among different species during evolution and might eventually contribute to unique evolutionary advantages by increasing tissue plasticity.
Methods
Raw sequence data and processing
Raw fastq files of ATAC-seq and RNA-seq data for five mouse tissues were gathered from ENCODE data portal (https://www.encodeproject.org/), including intestine, liver, lung, stomach, and kidney (Additional file 1) [39,40,41,42]. Two different development stages were considered in each tissue: embryonic day 14.5 (E14.5) and postnatal day 0 (P0) [39, 40]. The alignment bam files and narrowPeak bed files of H3K27ac for these five tissues with two development stages were also downloaded from ENCODE (Additional file 1).
ATAC-seq data of five tissues with two development stages were separately processed by AIAP package that contained an optimized ATAC-seq data QC and analysis pipeline with default parameters [93]. Open chromatin regions (OCR) generated by AIAP were used in downstream analysis. Then, the two replicates of peaks of same tissue from two different stages were combined by using mergeBed [94].
RNA-seq data of five tissues with two development stages were processed by Cutadapt (v2.7; --quality-cutoff = 15,10 --minimum-length = 36), FastQC (v0.11.4), and STAR (v2.5.2b; --quantMode TranscriptomeSAM --outWigType bedGraph --outWigNorm RPM) to do the trimming, QC report, and mouse genome mapping (mm10) [95,96,97]. Then, the mouse gene expression was calculated by featureCounts (-p -T 4 -Q 10) [98] based on GENCODE vM20 gene annotation. The expression (TPM) of CHIT1 and HAVCR1 in those five human tissues were obtained from “GTEx_Analysis_2017-06-05_v8_RNASeQCv1.1.9_gene_tpm.gct” file, which was processed and normalized by GTEx Portal (https://www.gtexportal.org/home/) [99].
The H3K27ac alignment (bam file) and narrow peaks (bed file) of five mouse tissues at two developmental stages were downloaded from ENCODE data portal. The bamtobed of bedtools was used to process bam file and generate fragment file (bed files), which were further used to calculate the signal density (CPM) under each peak by using the intersectBed (-wa -c) command. The high confident H3K27ac narrow peak (with H3K27ac signal CPM > 3) were combined to the union peak set by using mergeBed, and used for downstream analysis.
Transposable element annotation data and spatial correlation with peaks
The transposable elements (TEs) data of mouse (mm10) were obtained from the UCSC database (http://hgdownload.soe.ucsc.edu/goldenPath/mm10/database/) [100]. And the simple repeats, satellites, and TEs shorter than 100 bp were removed from this TE dataset.
The GenometriCorr (Genometric Correlation) is an R package that is used to calculate the spatial correlation of genome-wide interval datasets with the null hypothesis: interval sets are spatially independent [7, 43]. The correlated interval sets would follow a detectable pattern of the location, such as being consistently nearby or far away from each other in genomic coordinates, or preferentially overlapping. The permutation tests with 100 times were performed to look for TE enrichment in the open chromatin regions.
Identification of accessible TEs
The accessible TEs in five mouse tissues were separately identified by two methods: (1) accessible TEs were firstly identified by the overlapping between TE and OCR peak with intersectBed (-f 0.5 -F 0.2), at least 50% overlap required as a fraction of TE element and at least 20% overlap required as a fraction of peak; (2) we calculated the Tn5 insertions of remaining TEs from method 1. Genome-wide Tn5 insertion counts were generated by AIAP, and Tn5 insertion of each remaining TEs and OCR peaks of five tissues at two development were calculated by using intersectBed, and further were normalized to the insertion per kilobase per million insertions (IPKM) based on total insertions of libraries and length of TEs and OCR peak. The 25% quantile IPKM of OCR peaks in each library was used as cutoff (Additional file 5: Fig. S9), and remaining TEs from method 1 with an IPKM over the cutoff were considered as accessible TEs. Finally, the accessible TEs identified by the above two methods were combined and used for analysis in the study. The coordinates of accessible TEs can be found in Additional file 2.
The intersectBed [94] was used to determine the numbers of accessible TEs across genomic features (exons, first exons, introns, and intergenic regions), which were defined by using GENCODE vM20 gene annotation, in the five mouse tissues. Then, the enrichment ratio of accessible TEs in each genomic feature was calculated by dividing the number of accessible TEs in each genomic feature to the number of all TEs in each genomic feature (Additional file 4: Table S2; Additional file 5: Fig. S1).
ATAC-seq data of mouse embryonic stem cell was obtained from GEO (accession number: GSE94249) and processed by the AIAP package to generate OCR peaks (ESC peaks) representing the chromatin open status in early development stage [101]. The accessible TEs identified in five mouse tissues that can overlap to ESC OCR peaks were defined as open in this early development stage (Additional file 4: Table S5).
Subfamily enrichment of accessible TEs
To measure the subfamily enrichment of accessible TEs in each tissue, log enrichment ratio (LER) was calculated by the observed number of accessible TEs in the subfamily over the expected number of all TEs in the subfamily across open chromatin regions by the following formula:
The individual copy numbers in each TE subfamily were count by using TE annotation dataset described above. The enriched subfamily of accessible TEs was calculated and filtered with the following two criteria: (1) the number of individual TE copies in one subfamily should be greater than 10, and (2) the LER value should be greater than 3 (Additional file 4: Table S7).
GO analysis of tissue-specific accessible TEs
Accessible TEs that were only identified in one tissue was defined as tissue-specific accessible TEs. GO enrichment analysis of tissue-specific accessible TEs was performed by using GREAT (version 3.0.0) [102] with the following settings: (1) species assembly: mouse, NCBI build 38; (2) background regions: whole genome; (3) association rule: basal plus extension. Top 20 biological process terms were generated by GREAT with cutoffs of Binom FDR Q-Val < 0.05 and Hyper FDR Q-Val < 0.05 simultaneously (Additional file 6). GO enrichment analysis of accessible TEs constitutively identified in five tissues was also performed by using GREAT with the above settings.
Defining genes associated with accessible TE-derived TSS
Mouse gene annotation (Gencode.vM20.annotation.gtf) was obtained from GENCODE to define the relationship between genes and accessible TEs [103]. The 500 bp upstream of full-length genes defined as the gene’s promoter region were used to overlap with accessible TEs by intersectBed. TE-derived TSS was defined by the intersection between accessible TEs and furthest 5′ end transcription start sites (TSS) of genes. The gene-type distribution of accessible TEs-derived TSS across five tissues was generated based on Gencode.vM20.annotation.gtf file of mouse genes (Additional file 7). CAGE-TSS data from FANTOM5 were used to validate accessible TEs-derived TSS across five tissues (Additional file 7) [73]. The examples of accessible TE-derived TSS and CAGE TSS locations were visualized on WashU Epigenome Browser [104].
Differential accessible TEs and motif enrichment analysis
For each tissue, we calculated reads count table of peaks at E14.5 and P0 by using AIAP package and used edgeR to identify differential accessible regions (DARs) in the comparison between two development stages (abs (log2(Foldchange)) > 1 and FDR < 0.05, Additional file 5: FigureS10) [105, 106]. Then, the accessible TEs overlapped with DARs were identified as differential accessible TEs between two stages in five tissues: (1) those accessible TEs with positive foldchange were defined as P0day-specific TEs that were more open at P0; and (2) accessible TEs with negative foldchange were defined as E14.5day-specific TEs that were more open at E14.5. The rest of the accessible TEs did not show the difference of accessibility between two stages (Additional file 8: Table S1).
To explore the relation of differential accessible TEs on the gene expression at two stages, we assigned E14.5day-specific and P0day-specific TEs in five tissues to their nearest genes (the distance between TE and gene should be less than 20 kb), and the expression of those associated genes was tested by using the Wilcoxon signed-rank test to assess the significance of the difference between two development stages.
The enriched TFBS motifs of differential accessible TEs at E14.5 and P0 in five tissues were respectively discovered by using findMotifsGenome.pl (-size given) of HOMER software [107]. The enriched de novo motifs were selected with three conditions: (1) at least 10% of differential accessible TEs contained the motif; (2) match score of motifs should be greater than 0.9; (3) P value of the motif should be less than 1e−11. Then the known transcription factor genes (TF genes) with a motif match score > 0.9 to the enriched de novo motifs were extracted from HOMER results (Additional file 8).
Estimating the evolutionary conservation of accessible TEs
The evolutionary conservation of accessible TEs was measured by using phastCons conservation scores from mm10.60way.phastCons.bw file downloaded from UCSC, which contained conservation scores for alignments of 59 vertebrate genomes with mouse genome generated by phastCons program [100, 108]. The bigwig file was transferred to the bedGraph file with bigWigToBedGraph tool [109]. In total, 20,000 accessible TEs were randomly chosen and expanded 5 kb from the center at both sides, and their base-pair level phastCons scores were obtained by using intersectBed. Then, each 10-kb region was divided into 100-bp windows, and the averaged conservation scores of each 100-bp region were computed separately. Finally, the conservation scores at 100 bp resolution of all 20,000 accessible TEs were averaged and plotted. The evolutionary conservation of different genomic features, including exons, introns, and peaks without TEs, was separately analyzed by using the above method with 20,000 10-kb regions that were randomly chosen from each genomic feature.
Open chromatin regions with TE or without TE in five mouse tissues were aligned to rat (rn6) and human (hg38) genome by using liftOver software with “-minMatch = 0.6” [64]. The mouse open chromatin regions that can successfully lift over to the rat or human genome were defined as orthologous in rat or human genome. Meanwhile, to measure the orthologous conservation at the genome-wide level, the whole mouse genome was first divided into 200-bp windows, and the orthologous regions were defined as the same standard as orthologous OCR described above, as well as those 200-bp windows that did not overlap with TEs. The orthologous conservation was also measured for the whole TEs in the mouse genome (Additional file 9: Table S1).
Ortholog protein-coding genes with TE-derived TSS in three species (mouse, rat, and human) were first determined by using ortholog information obtained from Ensembl [110]. The mouse-specific and mouse-rat ortholog genes were further validated by using blastp, the protein sequence of mouse genes against rat (taxid: 10116) and human (taxid: 9606) reference database on NCBI, with the cutoff of both coverage and identify > 60% [111].
To assess the conservation of lincRNA, the cDNA sequence of lincRNAs were obtained from “Long non-coding RNA gene annotation” file of GENCODE (vM21 version), and then blastn method with “somewhat similar sequences” parameter was used to search each cDNA against rat (taxid: 10116) and human (taxid: 9606) reference RNA sequences [103, 112]. The orthologous lincRNA was defined as both the identity and coverage of the top blast hit to rat, or human reference RNA sequences can be over 60% (Additional file 9).
Conservation of the Timd2 gene
Timd2 and Havcr1 genes of mouse were identified as duplicated genes in the Duplicated Genes Database (DGD) [113]. The sequence of cDNA and protein between Timd2 and Havcr1 was obtained from the Ensembl database (https://useast.ensembl.org/Mus_musculus), and the pair-wise alignment results were generated by Mview (Additional file 5: Fig. S3) [110, 114]. The gene cDNA sequences of Havcr1, Timd2, and Timd4 in mouse and rat were downloaded from the Ensembl database, multiple sequence alignment was performed by using muscle [115]. Then MEGA was used to construct the evolutionary tree of those genes with the neighbor-joining method [116].
The exon sequences of mouse Havcr1 and Timd2 genes were downloaded from the Ensembl database [117]. Exons of Havcr1 were aligned to the Timd2 sequence by using blastn with “somewhat similar sequences” parameter to identify the homologous exons between two genes [112]. The exon sequence coverage, identity, and gaps between homologous of Havcr1 and Timd2 were estimated based on the blastn results (Additional file 10).
FIMO software was used to identify the TFBS motifs in the RLTR14-int elements based on motif weigh matrix file (JASPAR_CORE_2016_vertebrates.meme) from JASPAR [118, 119]. The ChIP-seq signals of Hnf4a, Rxra, Cebpb, and Pol2 genes were collected from GEO (GSM2406338, GSM1163178, GSM1854433, and GSM864688) and displayed on WashU Epigenome Browser [101, 104, 120].
The consensus sequence of RLTR14-int was downloaded from Repbase and was used to perform the pair-wise alignment with all RLTR14-int individual copies in the mouse genome by using muscle [115, 121]. The genomic information of proteins within the RLTR14-int consensus sequence was obtained from Dfam database [122]. FIMO software was applied to identify TFBS motifs for each RLTR14-int copy that can align to Timd2-TSS RLTR14-int element [119]. The base-pair conservation score of RLTR14-int alignments was calculated by the following formula:
Transcripts assemble
The mouse gene transcripts of five tissues at two development stages were assembled based on RNA-seq data. First, the sorted BAM files were generated after quality trimming by Cutadapt (v2.7) with parameter “-quality-cutoff = 15,10 --minimum-length = 36” and reads mapping to mouse genome (mm10) by STAR (version 2.5.2b) with the parameter “--outSAMtype BAM SortedByCoordinate --outFilterMultimapNmax 20 --outSAMattributes NH HI NM MD AS XS --twopassMode Basic --outFilterType BySJout --limitBAMsortRAM 100000000000” [95, 97]. Then, StringTie (v2.0.4) with the parameter “-f 0.15 -a 20 -j 10 -m 200” was applied to assemble transcripts based on sorted BAM files [123]. Next, StringTie took as the input of a list of transcripts GTF files with coverage of at least 10 and merged these transcripts into a non-redundant set and compared assembled transepts to GENCODE gene annotation vM20. The quantification of the unified set of transcripts was calculated by RSEM (v1.3.0; -p 12 --bam) separately for five tissues at two stages [124].
In total, 316 TE-derived TSS genes were found to express in at least one tissue (mean expression FPKM > 0.1) and were classified into three groups based on the TSS usage of expressed transcripts: (1) only 5′ end TE-derived TSS used; (2) none of 5′ end TE-derived TSS used; and (3) both 5′ end TE-derived TSS and non-TE TSS used (Additional file 11: Table S1). Then, the expression percentage of TE-derived transcripts in each gene was calculated by the sum of the expression of TE-derived transcripts dividing the total gene expression. Based on the RNA-seq alignment (bam files) and gene GFF annotation file, the sashimi plots of each gene were generated by using sashimi_plot command in MISO framework to visualize the RNA-seq read densities along with exons and junctions, as well as visualize the structure of the gene’s isoforms [125, 126].
CAGE-seq peak-based expression table of robust CAGE-seq peaks for mouse samples with annotation was downloaded from FANTOM5 project [73]. The expression table of CAGE peaks at the embryo development stage of the intestine, liver, lung, stomach, and kidney tissues was downloaded from the FANTOM data portal (https://fantom.gsc.riken.jp/data/) and further extracted by in-house script. The CAGE-seq peaks were assigned to either TE-derived TSS or non-TE TSS by using intersectBed with 1 bp overlap. Then the contribution of TE-derived TSS was calculated based on the expression level of CAGE-seq peak associated with TE-derived TSS in the total expression level of CAGE-seq peaks associated with both TE-derived TSS and non-TE TSS.
Availability of data and materials
The ATAC-seq, RNA-seq, and H3k27ac data for 5 mouse tissues that were processed and analyzed in current study are available from ENCODE data portal, and their accession numbers were recorded in Additional file 1 [41, 42]. The CAGE data are available from FANTOM5 project [73]. ATAC-seq data of mouse embryonic stem cell is available from GEO (GSE94249) [101]. The ChIP-seq datasets are available from GEO (GSM2406338, GSM1163178, GSM1854433, and GSM864688) [101].
References
International Human Genome Sequencing C. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–45.
Genomes Project C, Abecasis GR, Auton A, Brooks LD, MA DP, Durbin RM, Handsaker RE, Kang HM, Marth GT, GA MV. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65.
Alexander RP, Fang G, Rozowsky J, Snyder M, Gerstein MB. Annotating non-coding regions of the genome. Nat Rev Genet. 2010;11:559–71.
Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–61.
Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, Ng HH, Bourque G. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 2010;42:631–4.
Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921.
Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet. 2013;9:e1003470.
Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9:397–405.
Jacques PE, Jeyakani J, Bourque G. The majority of primate-specific regulatory sequences are derived from transposable elements. PLoS Genet. 2013;9:e1003504.
Ito J, Sugimoto R, Nakaoka H, Yamada S, Kimura T, Hayano T, Inoue I. Systematic identification and characterization of regulatory elements derived from human endogenous retroviruses. PLoS Genet. 2017;13:e1006883.
Chuong EB, Elde NC, Feschotte C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science. 2016;351:1083–7.
Hollister JD, Gaut BS. Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 2009;19:1419–28.
Zhang W, Fan Z, Han E, Hou R, Zhang L, Galaverni M, Huang J, Liu H, Silva P, Li P. Hypoxia adaptations in the grey wolf (Canis lupus chanco) from Qinghai-Tibet plateau. PLoS Genet. 2014;10:e1004466.
Xie M, Hong C, Zhang B, Lowdon RF, Xing X, Li D, Zhou X, Lee HJ, Maire CL, Ligon KL, et al. DNA hypomethylation within specific transposable element families associates with tissue-specific enhancer landscape. Nat Genet. 2013;45:836–41.
Lynch VJ, Leclerc RD, May G, Wagner GP. Transposon-mediated rewiring of gene regulatory networks contributed to the evolution of pregnancy in mammals. Nat Genet. 2011;43:1154–9.
Bourque G, Leong B, Vega VB, Chen X, Lee YL, Srinivasan KG, Chew JL, Ruan Y, Wei CL, Ng HH, Liu ET. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18:1752–62.
Jang HS, Shah NM, Du AY, Dailey ZZ, Pehrsson EC, Godoy PM, Zhang D, Li D, Xing X, Kim S, et al. Transposable elements drive widespread expression of oncogenes in human cancers. Nat Genet. 2019;51:611–7.
van de Lagemaat LN, Landry JR, Mager DL, Medstrand P. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet. 2003;19:530–6.
Romanish MT, Lock WM, van de Lagemaat LN, Dunn CA, Mager DL. Repeated recruitment of LTR retrotransposons as promoters by the anti-apoptotic locus NAIP during mammalian evolution. PLoS Genet. 2007;3:e10.
Romanish MT, Nakamura H, Lai CB, Wang Y, Mager DL. A novel protein isoform of the multicopy human NAIP gene derives from intragenic Alu SINE promoters. PLoS One. 2009;4:e5761.
Ekram MB, Kang K, Kim H, Kim J. Retrotransposons as a major source of epigenetic variations in the mammalian genome. Epigenetics. 2012;7:370–82.
Farshchian M, Matin MM, Armant O, Geerts D, Dastpak M, Nakhaei-Rad S, Tajeran M, Jebelli A, Shahriyari M, Bahrami M. Suppression of dsRNA response genes and innate immunity following Oct4, Stella, and Nanos2 overexpression in mouse embryonic fibroblasts. Cytokine. 2018;106:1–11.
Kazachenka A,Bertozzi T, Sjoberg-Herrera M, Walker N, Gardner J, Gunning R, Pahita E, Adams S, Adams D, Ferguson-Smith A. Identification, Characterization, and Heritability of Murine Metastable Epialleles: Implications for Non-genetic Inheritance. Cell. 2018; 175(5): 1259–1271.e13.
Chishima T, Iwakiri J, Hamada M. Identification of transposable elements contributing to tissue-specific expression of long non-coding RNAs. Genes (Basel). 2018;9(1):23.
Franke V, Ganesh S, Karlic R, Malik R, Pasulka J, Horvat F, Kuzman M, Fulka H, Cernohorska M, Urbanova J, et al. Long terminal repeats power evolution of genes and gene expression programs in mammalian oocytes and zygotes. Genome Res. 2017;27:1384–94.
Bogutz AB, Brind'Amour J, Kobayashi H, Jensen KN, Nakabayashi K, Imai H, Lorincz MC, Lefebvre L. Evolution of imprinting via lineage-specific insertion of retroviral promoters. Nat Commun. 2019;10:5674.
Robbez-Masson L, Tie CHC, Conde L, Tunbak H, Husovsky C, Tchasovnikarova IA, Timms RT, Herrero J, Lehner PJ, Rowe HM. The HUSH complex cooperates with TRIM28 to repress young retrotransposons and new genes. Genome Res. 2018;28:836–45.
Todd CD, Deniz O, Taylor D, Branco MR. Functional evaluation of transposable elements as enhancers in mouse embryonic and trophoblast stem cells. Elife. 2019;8:e44344.
Thompson PJ, Macfarlan TS, Lorincz MC. Long terminal repeats: from parasitic elements to building blocks of the transcriptional regulatory repertoire. Mol Cell. 2016;62:766–76.
Pasquesi GIM, Perry BW, Vandewege MW, Ruggiero RP, Schield DR, Castoe TA. Vertebrate lineages exhibit diverse patterns of transposable element regulation and expression across tissues. Genome Biol Evol. 2020;12:506–21.
Pavlicev M, Hiratsuka K, Swaggart KA, Dunn C, Muglia L. Detecting endogenous retrovirus-driven tissue-specific gene transcription. Genome Biol Evol. 2015;7:1082–97.
Rebollo R, Miceli-Royer K, Zhang Y, Farivar S, Gagnier L, Mager DL. Epigenetic interplay between mouse endogenous retroviruses and host genes. Genome Biol. 2012;13:R89.
Reiss D, Zhang Y, Rouhi A, Reuter M, Mager DL. Variable DNA methylation of transposable elements: the case study of mouse early transposons. Epigenetics. 2010;5:68–79.
Sundaram V, Cheng Y, Ma Z, Li D, Xing X, Edge P, Snyder MP, Wang T. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 2014;24:1963–76.
Brocks D, Schmidt CR, Daskalakis M, Jang HS, Shah NM, Li D, Li J, Zhang B, Hou Y, Laudato S, et al. DNMT and HDAC inhibitors induce cryptic transcription start sites encoded in long terminal repeats. Nat Genet. 2017;49:1052–60.
Nagarajan RP, Zhang B, Bell RJ, Johnson BE, Olshen AB, Sundaram V, Li D, Graham AE, Diaz A, Fouse SD, et al. Recurrent epimutations activate gene body promoters in primary glioblastoma. Genome Res. 2014;24:761–74.
Sun W, Samimi H, Gamez M, Zare H, Frost B. Pathogenic tau-induced piRNA depletion promotes neuronal death through transposable element dysregulation in neurodegenerative tauopathies. Nat Neurosci. 2018;21:1038–48.
Kelly M, Lihua S, Zhe Z, Li S, Yoselin P, Michelle P, Sullivan Kathleen E. Transposable element dysregulation in systemic lupus erythematosus and regulation by histone conformation and Hsp90. Clin Immunol. 2018;197:6–18.
Gorkin DU, Barozzi I, Zhao Y, Zhang Y, Huang H, Lee AY, Li B, Chiou J, Wildberg A, Ding B. An atlas of dynamic chromatin landscapes in mouse fetal development. Nature. 2020;583:744–51.
He P, Williams BA, Trout D, Marinov GK, Amrhein H, Berghella L, Goh S-T, Plajzer-Frick I, Afzal V, Pennacchio LA. The changing mouse embryo transcriptome at whole tissue and single-cell resolution. Nature. 2020;583:760–7.
Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74.
Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, Hilton JA, Jain K, Baymuradov UK, Narayanan AK. The encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018;46:D794–801.
Favorov A, Mularoni L, Cope LM, Medvedeva Y, Mironov AA, Makeev VJ, Wheelan SJ. Exploring massive, genome scale datasets with the GenometriCorr package. PLoS Comput Biol. 2012;8:e1002529.
Pehrsson EC, Choudhary MN, Sundaram V, Wang T. The epigenomic landscape of transposable elements across normal human development and anatomy. Nat Commun. 2019;10:1–16.
Sundaram V, Wang T. Transposable element mediated innovation in gene regulatory landscapes of cells: re-visiting the “gene-battery” model. BioEssays. 2018;40:1700155.
Lv Y, Hu F, Zhou Y, Wu F, Gaut BS. Maize transposable elements contribute to long non-coding RNAs that are regulatory hubs for abiotic stress response. BMC Genomics. 2019;20:864.
Wang Z, Schwacke R, Kunze R. DNA damage-induced transcription of transposable elements and long non-coding RNAs in arabidopsis is rare and ATM-dependent. Mol Plant. 2016;9:1142–55.
Hutchins AP, Pei D. Transposable elements at the center of the crossroads between embryogenesis, embryonic stem cells, reprogramming, and long non-coding RNAs. Sci Bull (Beijing). 2015;60:1722–33.
Kannan S, Chernikova D, Rogozin IB, Poliakov E, Managadze D, Koonin EV, Milanesi L. Transposable element insertions in long intergenic non-coding RNA genes. Front Bioeng Biotechnol. 2015;3:71.
Hadjiargyrou M, Delihas N. The intertwining of transposable elements and non-coding RNAs. Int J Mol Sci. 2013;14:13307–28.
Papin C, Simonelig M. Embryonic development is controlled by small non-coding RNAs derived from transposable elements. Med Sci (Paris). 2011;27:1050–2.
Trizzino M, Kapusta A, Brown CD. Transposable elements generate regulatory novelty in a tissue-specific fashion. BMC Genomics. 2018;19:468.
Jin P, Qin S, Chen X, Song Y, Li-Ling J, Xu X, Ma F. Evolutionary rate of human tissue-specific genes are related with transposable element insertions. Genetica. 2012;140:513–23.
Donlin MJ, Lisch D, Freeling M. Tissue-specific accumulation of MURB, a protein encoded by MuDR, the autonomous regulator of the Mutator transposable element family. Plant Cell. 1995;7:1989–2000.
Trizzino M, Park Y, Holsbach-Beltrame M, Aracena K, Mika K, Caliskan M, Perry GH, Lynch VJ, Brown CD. Transposable elements are the primary source of novelty in primate gene regulation. Genome Res. 2017;27:1623–33.
Yan F, Di S, Takahashi R. CACTA-superfamily transposable element is inserted in MYB transcription factor gene of soybean line producing variegated seeds. Genome. 2015;58:365–74.
Sundaram V, Choudhary MN, Pehrsson E, Xing X, Fiore C, Pandey M, Maricque B, Udawatta M, Ngo D, Chen Y, et al. Functional cis-regulatory modules encoded by mouse-specific endogenous retrovirus. Nat Commun. 2017;8:14550.
Geraud C, Koch PS, Zierow J, Klapproth K, Busch K, Olsavszky V, Leibing T, Demory A, Ulbrich F, Diett M, et al. GATA4-dependent organ-specific endothelial differentiation controls liver development and embryonic hematopoiesis. J Clin Invest. 2017;127:1099–114.
Borok MJ, Papaioannou VE, Sussel L. Unique functions of Gata4 in mouse liver induction and heart development. Dev Biol. 2016;410:213–22.
Watt AJ, Zhao R, Li J, Duncan SA. Development of the mammalian liver and ventral pancreas is dependent on GATA4. BMC Dev Biol. 2007;7:37.
Zhao R, Watt AJ, Li J, Luebke-Wheeler J, Morrisey EE, Duncan SA. GATA6 is essential for embryonic development of the liver but dispensable for early heart formation. Mol Cell Biol. 2005;25:2622–31.
Bosse T, Piaseckyj CM, Burghard E, Fialkovich JJ, Rajagopal S, Pu WT, Krasinski SD. Gata4 is essential for the maintenance of jejunal-ileal identities in the adult mouse small intestine. Mol Cell Biol. 2006;26:9060–70.
Koutsourakis M, Langeveld A, Patient R, Beddington R, Grosveld F. The transcription factor GATA6 is essential for early extraembryonic development. Development. 1999;126:723–32.
Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, Furey TS, Harte RA, Hsu F. The UCSC genome browser database: update 2006. Nucleic Acids Res. 2006;34:D590–8.
Chuong EB, Elde NC, Feschotte C. Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet. 2017;18:71.
Hou J, Lu D, Mason AS, Li B, Xiao M, An S, Fu D. Non-coding RNAs and transposable elements in plant genomes: emergence, regulatory mechanisms and roles in plant development and stress responses. Planta. 2019;250:23–40.
Morata J, Marin F, Payet J, Casacuberta JM. Plant lineage-specific amplification of transcription factor binding motifs by miniature inverted-repeat transposable elements (MITEs). Genome Biol Evol. 2018;10:1210–20.
Kanneganti M, Kamba A, Mizoguchi E. Role of chitotriosidase (chitinase 1) under normal and disease conditions. J Epithel Biol Pharmacol. 2012;5:1–9.
Elmonem MA, van den Heuvel LP, Levtchenko EN. Immunomodulatory effects of Chitotriosidase enzyme. Enzyme Res. 2016;2016:2682680.
Schyns J, Bureau F, Marichal T. Lung interstitial macrophages: past, present, and future. J Immunol Res. 2018;2018:5160794.
Cuadros T, Trilla E, Vila MR, de Torres I, Vilardell J, Messaoud NB, Salcedo M, Sarro E, Lopez-Hellin J, Blanco A, et al. Hepatitis a virus cellular receptor 1/kidney injury molecule-1 is a susceptibility gene for clear cell renal cell carcinoma and hepatitis A virus cellular receptor/kidney injury molecule-1 ectodomain shedding a predictive biomarker of tumour progression. Eur J Cancer. 2013;49:2034–47.
Wang S, Xiao X, Zhou X, Huang T, Du C, Yu N, Mo Y, Lin L, Zhang J, Ma N. TFPI-2 is a putative tumor suppressor gene frequently inactivated by promoter hypermethylation in nasopharyngeal carcinoma. BMC Cancer. 2010;10:617.
Noguchi S, Arakawa T, Fukuda S, Furuno M, Hasegawa A, Hori F, Ishikawa-Kato S, Kaida K, Kaiho A, Kanamori-Katayama M. FANTOM5 CAGE profiles of human and mouse samples. Scientific data. 2017;4:170112.
Clayton EA, Rishishwar L, Huang T-C, Gulati S, Ban D, McDonald JF, Jordan IK. An atlas of transposable element-derived alternative splicing in cancer. Philos Trans R Soc B. 2020;375:20190342.
Cowley M, Oakey RJ. Transposable elements re-wire and fine-tune the transcriptome. PLoS Genet. 2013;9:e1003234.
Lawson T, Blatt MR. Stomatal size, speed, and responsiveness impact on photosynthesis and water use efficiency. Plant Physiol. 2014;164:1556–70.
Niven JE, Laughlin SB. Energy limitation as a selective pressure on the evolution of sensory systems. J Exp Biol. 2008;211:1792–804.
Blekhman R, Oshlack A, Chabot AE, Smyth GK, Gilad Y. Gene regulation in primates evolves under tissue-specific selection pressures. PLoS Genet. 2008;4:e1000271.
Gerdes P, Richardson SR, Mager DL, Faulkner GJ. Transposable elements in the mammalian embryo: pioneers surviving through stealth and service. Genome Biol. 2016;17:100.
Zhang B, Zhou Y, Lin N, Lowdon RF, Hong C, Nagarajan RP, Cheng JB, Li D, Stevens M, Lee HJ, et al. Functional DNA methylation differences between tissues, cell types, and across individuals discovered using the M&M algorithm. Genome Res. 2013;23:1522–40.
Conley AB, Jordan IK. Identification of transcription factor binding sites derived from transposable element sequences using ChIP-seq. Methods Mol Biol. 2010;674:225–40.
Taube JH, Allton K, Duncan SA, Shen L, Barton MC. Foxa1 functions as a pioneer transcription factor at transposable elements to activate Afp during differentiation of embryonic stem cells. J Biol Chem. 2010;285:16135–44.
Becker KG, Swergold GD, Ozato K, Thayer RE. Binding of the ubiquitous nuclear transcription factor YY1 to a cis regulatory sequence in the human LINE-1 transposable element. Hum Mol Genet. 1993;2:1697–702.
Philpott C, Tovell H, Frayling IM, Cooper DN, Upadhyaya M. The NF1 somatic mutational landscape in sporadic human cancers. Hum Genomics. 2017;11:13.
Yap YS, McPherson JR, Ong CK, Rozen SG, Teh BT, Lee AS, Callen DF. The NF1 gene revisited - from bench to bedside. Oncotarget. 2014;5:5873–92.
Herriges M, Morrisey EE. Lung development: orchestrating the generation and regeneration of a complex organ. Development. 2014;141:502–13.
Morrisey EE, Hogan BL. Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev Cell. 2010;18:8–23.
Arner E, Daub CO, Vitting-Seerup K, Andersson R, Lilje B, Drablos F, Lennartsson A, Ronnerblad M, Hrydziuszko O, Vitezic M, et al. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science. 2015;347:1010–4.
Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–40.
Ecco G, Cassano M, Kauzlaric A, Duc J, Coluccio A, Offner S, Imbeault M, Rowe HM, Turelli P, Trono D. Transposable elements and their KRAB-ZFP controllers regulate gene expression in adult tissues. Dev Cell. 2016;36:611–23.
Ricci M, Peona V, Guichard E, Taccioli C, Boattini A. Transposable elements activity is positively related to rate of speciation in mammals. J Mol Evol. 2018;86:303–10.
Mersch B, Sela N, Ast G, Suhai S, Hotz-Wagenblatt A. SERpredict: detection of tissue- or tumor-specific isoforms generated through exonization of transposable elements. BMC Genet. 2007;8:78.
Liu S, Lyu C, Gontarz P, Miao B, Madden P, Wang T, Zhang B: Improving ATAC-seq data analysis with AIAP, a quality control and integrative analysis package. bioRxiv 686808. 2019. https://doi.org/10.1101/686808.
Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2.
Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–2.
Andrews S. FastQC: a quality control tool for high throughput sequence data. Cambridge: Babraham Bioinformatics, Babraham Institute; 2010.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30.
Consortium G. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–60.
Haeussler M, Zweig AS, Tyner C, Speir ML, Rosenbloom KR, Raney BJ, Lee CM, Lee BT, Hinrichs AS, Gonzalez JN. The UCSC genome browser database: 2019 update. Nucleic Acids Res. 2019;47:D853–8.
Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10.
McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495.
Frankish A, Diekhans M, Ferreira A-M, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47:D766–73.
Zhou X, Wang T. Using the Wash U Epigenome Browser to examine genome-wide sequencing data. Curr Protoc Bioinformatics. 2012;40:10.10. 11–4.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
Gontarz P, Fu S, Xing X, Liu S, Miao B, Bazylianska V, Sharma A, Madden P, Cates K, Yoo A, et al. Comparison of differential accessibility analysis strategies for ATAC-seq data. Sci Rep. 2020;10:10150.
Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89.
Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–50.
Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010;26:2204–7.
Yates AD, Achuthan P, Akanni W, Allen J, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, Azov AG, Bennett R. Ensembl 2020. Nucleic Acids Res. 2020;48:D682–8.
Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2007;36:D13–21.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10.
Ouedraogo M, Bettembourg C, Bretaudeau A, Sallou O, Diot C, Demeure O, Lecerf F. The duplicated genes database: identification and functional annotation of co-localised duplicated genes across genomes. PLoS One. 2012;7(11): e50653.
Brown NP, Leroy C, Sander C. MView: a web-compatible database search or multiple alignment viewer. Bioinformatics (Oxford). 1998;14:380–1.
Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7.
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4.
Chen Y, Cunningham F, Rios D, McLaren WM, Smith J, Pritchard B, Spudich GM, Brent S, Kulesha E, Marin-Garcia P. Ensembl variation resources. BMC Genomics. 2010;11:293.
Mathelier A, Fornes O, Arenillas DJ, Chen C-Y, Denay G, Lee J, Shi W, Shyr C, Tan G, Worsley-Hunt R. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2016;44:D110–5.
Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–8.
Zheng R, Wan C, Mei S, Qin Q, Wu Q, Sun H, Chen C-H, Brown M, Zhang X, Meyer CA. Cistrome Data Browser: expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Res. 2019;47:D729–35.
Bao W, Kojima KK, Kohany O. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 2015;6:11.
Hubley R, Finn RD, Clements J, Eddy SR, Jones TA, Bao W, Smit AF, Wheeler TJ. The Dfam database of repetitive DNA families. Nucleic Acids Res. 2016;44:D81–9.
Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–5.
Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323.
Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–15.
Katz Y, Wang ET, Silterra J, Schwartz S, Wong B, Thorvaldsdóttir H, Robinson JT, Mesirov JP, Airoldi EM, Burge CB. Quantitative visualization of alternative exon expression from RNA-seq data. Bioinformatics. 2015;31:2400–2.
Review history
The review history is available as Additional file 12.
Peer review information
Yixin Yao was the primary editor of this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Authors’ information
Twitter handles: @twang5 (Ting Wang); Bo_Zhang_WashU (Bo Zhang).
Funding
This work was supported by the National Institutes of Health [U24ES026699, U01HG009391, and R25DA027995], the Goldman Sachs Philanthropy Fund [Emerson Collective], and the Chan Zuckerberg Initiative [Human Cell Atlas Seed Network]. Funding for open access charge: National Institutes of Health.
Author information
Authors and Affiliations
Contributions
BZ and TW designed and supervised the study. BM, SF, CL, and PG processed data and performed analysis. BM, TW, and BZ wrote the manuscript with input from all the other authors. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1.
The epigenomic data of five tissues at embryonic day 14.5 and postnatal day 0 development stages from the ENCODE.
Additional file 2.
The accessible TEs identified in the five tissues.
Additional file 3.
The spatial correlation between open chromatin regions and transposable elements (TEs) in five tissues.
Additional file 4: Table S1.
The total number of peaks, H3k27ac regions, and accessible TEs in five tissues. Table S2. The genomic distribution of accessible TEs in 5 mouse tissues. Table S3. The number of accessible TEs shared among the five tissues. Table S4. Enriched TF binding motifs identified in TEs commonly accessible in five tissues. Table S5. The accessibility of TEs in mouse embryonic stem cells. Table S6. The class distribution of accessible TEs in five tissues. Table S7. The subfamily enrichment of those accessible TEs.
Additional file 5: Fig. S1.
Genomic distribution of accessible TEs of 5 mouse tissues in the mouse genome. Fig. S2. Epigenome browser view of ATAC-seq and RNA-seq signals of Chit1 gene. Fig. S3. Pair-wise alignment of cDNA and protein sequences between Timd2 and Havcr1 genes. Fig. S4. Epigenome browser view of ATAC-seq and RNA-seq signals of Timd2 gene. Fig. S5. Motif analysis of RLTR14-int elements. Fig. S6. Sashimi plot for the transcripts of genes at two development stages of five tissues. Fig. S7. The TE-derived TSS genes with transcript start site overlapped with CAGE Peaks. Fig. S8. Pair-wise alignment between the DNA sequences around the TSS of mouse Timd2 and Havcr1 genes. Fig. S9. The distribution of Tn5 insertion number in TE and Peak at E14.5 and P0 development stages of 5 tissues. Fig. S10. Percentage of dynamically changed Peaks between E14.5 and P0 in five mouse tissues.
Additional file 6.
The enriched GO biology process items of accessible TEs that only identified in one tissue.
Additional file 7: Table S1.
The number of accessible TEs located in the gene’s promoter and contributed to the TSS of genes. Table S2. The annotation of accessible TE that derived TSS of genes in five tissue. Table S3. Examples of accessible TEs that derived TSS.
Additional file 8: Table S1.
The dynamic changes of accessible TEs between two development stages of five tissues. Table S2. The number of stage-specific accessible TEs located in the gene’s promoter regions. Table S3-S7. The enriched TF binding motifs of stage-specific accessible TEs in intestine, liver, lung, stomach, and kidney. Table S8. Expression of TF genes at two development stages of five tissues.
Additional file 9: Table S1.
The evolutionary conservation of open chromatin regions with TEs or without TEs across five mouse tissues by comparing with the human and rat genome. Table S2. Conservation of accessible TEs that contained the TSSs of genes in mouse, rat, and human. Table S3. The ortholog of accessible TE-derived TSS genes in mouse, rat, and human. Table S4. The ortholog of accessible TE-derived lincRNAs in three species.
Additional file 10.
The percentage of identity, coverage, and gaps for the exon’s alignment between Timd2 and Havcr1 genes.
Additional file 11: Table S1.
The number of genes that their transcripts used 5′ end TE-derived TSS and non-TE derived TSS. Table S2. The usage of TE-derived TSS in the transcripts of genes overlapped CAGE peaks.
Additional file 12.
Review history.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Miao, B., Fu, S., Lyu, C. et al. Tissue-specific usage of transposable element-derived promoters in mouse development. Genome Biol 21, 255 (2020). https://doi.org/10.1186/s13059-020-02164-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13059-020-02164-3