- Method
- Open access
- Published:
pipeComp, a general framework for the evaluation of computational pipelines, reveals performant single cell RNA-seq preprocessing tools
Genome Biology volume 21, Article number: 227 (2020)
Abstract
We present pipeComp (https://github.com/plger/pipeComp), a flexible R framework for pipeline comparison handling interactions between analysis steps and relying on multi-level evaluation metrics. We apply it to the benchmark of single-cell RNA-sequencing analysis pipelines using simulated and real datasets with known cell identities, covering common methods of filtering, doublet detection, normalization, feature selection, denoising, dimensionality reduction, and clustering. pipeComp can easily integrate any other step, tool, or evaluation metric, allowing extensible benchmarks and easy applications to other fields, as we demonstrate through a study of the impact of removal of unwanted variation on differential expression analysis.
Background
Single-cell RNA-sequencing (scRNAseq) and the set of attached analysis methods are evolving fast, with more than 560 software tools available to the community [1], roughly half of which are dedicated to tasks related to data processing such as clustering, ordering, dimension reduction, or normalization. This increase in the number of available tools follows the development of new sequencing technologies and the growing number of reported cells, genes, and cell populations [2]. As data processing is a critical step in any scRNAseq analysis, affecting downstream analysis and interpretation, it is critical to evaluate the available tools.
A number of good comparison and benchmark studies have already been performed on various steps related to scRNAseq processing and analysis and can guide the choice of methodology [3–21]. However, these recommendations need constant updating and often leave open many details of an analysis. Another missing aspect of current benchmarking studies is their limitation to capture all aspects of a scRNAseq processing workflow. Although previous benchmarks already brought valuable recommendations for data processing, some only focused on one aspect of data processing (e.g., [14]), did not evaluate how the tool selection affects downstream analysis (e.g., [17]) or did not tackle all aspects of data processing, such as doublet identification or cell filtering (e.g., [18]). A thorough evaluation of the tools covering all major processing steps is however urgently needed as previous benchmarking studies highlighted that a combination of tools can have a drastic impact on downstream analysis, such as differential expression analysis and cell-type deconvolution[3, 18]. It is then critical to evaluate not only the single effect of a preprocessing method but also its positive or negative interaction with all parts of a workflow.
Here, we develop a flexible R framework for pipeline comparison and apply it to the evaluation of the various steps of analysis leading from an initial count matrix to a cluster assignment, which are critical in a wide range of applications. We collected real datasets of known cell composition (Table 1) and used a variety of evaluation metrics to investigate in a multilevel fashion the impact of various parameters and variations around a core scRNAseq pipeline. Although we use some datasets based on other protocols, our focus is especially on droplet-based datasets that do not include exogenous control RNA (i.e., spike-ins); see Table 1 and Additional File 1: Figure S1 for more details. In addition to previously used benchmark datasets with true cell labels [6, 15], we simulated two datasets with a hierarchical subpopulation structure based on real 10x human and mouse data using muscat [22]. Since graph-based clustering [23] was previously shown to consistently perform well across several datasets [6, 15], we used the Seurat pipeline as the starting point to perform an integrated investigation of (1) doublet identification, (2) cell filtering, (3) normalization, (4) feature selection, (5) dimension reduction, and (6) clustering. We compared competing approaches and also explored more fine-grained parameters and variations on common methods. Importantly, the success of methods at a certain analytical step might be dependent on choices at other steps. Therefore, instead of evaluating each step in isolation, we developed a general framework for evaluating nested variations on a pipeline and suggest a multilevel panel of metrics. Finally, we evaluate several recent methods and provide practical recommendations.
Results
A flexible framework for pipeline evaluation
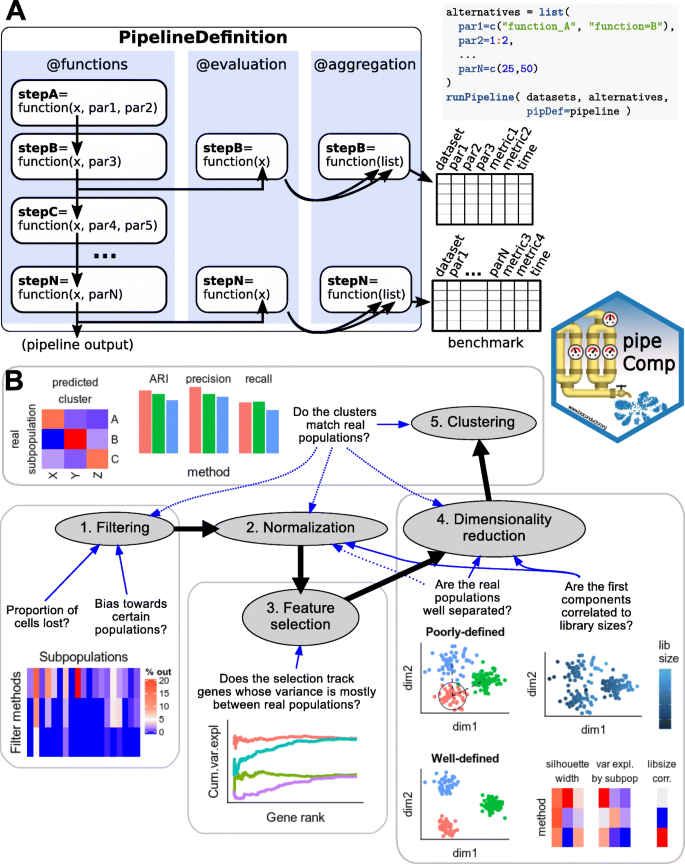
The pipeComp package defines a pipeline as, minimally, a list of functions executed consecutively on the output of the previous one (Fig. 1a; see also Additional File 2). In addition, optional benchmark functions can be set for each step to provide standardized, multi-layered evaluation metrics. Given such a PipelineDefinition object, a set of alternative parameters (which might include different subroutines calling different methods) and benchmark datasets, the runPipeline function then proceeds through all combinations of arguments (or a desired subset), avoiding recomputing the same step twice without the need to save all alternative intermediates, and compiling evaluations (including run time) on the fly (see Additional File 2 for more details). Variations in a given parameter can be evaluated using all metrics from this point downward in the pipeline. This is especially important because end-point metrics, such as the adjusted Rand index (ARI) [24] for clustering, are not perfect. For example, although the meaning of an ARI score is independent of the number of true subpopulations [25], the number of clusters called is by far the most important determinant of the score: the farther it is from the actual number of subpopulations, the worse the ARI (Additional File 1: Figure S2–3). In this context, one strategy has been to cluster across various resolutions and only consider the results that have the right number of clusters [6]. While this has the virtue of making the results comparable, in practice, the number of subpopulations is typically unknown and tools that operate well in this optimal context might not necessarily be best overall. Clustering results are also very sensitive and might not always capture improvements in earlier steps of the pipeline. In addition, we wanted to assess whether the effect of a given parameter alteration is robust to changes in the rest of the pipeline. We therefore monitored several complementary metrics across multiple steps of the process. We proceeded in a step-wise fashion, first testing a large variety of parameters at the early steps of the pipeline along with only a set of mainstream options downstream, then selecting main alternatives and proceeding to a more detailed benchmark of the next step (Fig. 1b).
Overview of the pipeComp framework and its application to a scRNAseq clustering pipeline. a The package is built around a PipelineDefinition class which defines a set of functions to be executed consecutively, as well as optional evaluation functions for each step. Each step can accept a number of parameters whose alternative values are provided as a list. All (or subsets of) combinations of parameters can then be simultaneously ran and evaluated using the runPipeline function (see Additional File 2 for more details). b Scheme representing the application of pipeComp to evaluate a range of methods that are commonly used in scRNAseq studies and some of the metrics monitored at various steps
Filtering
Doublet detection
Doublets, defined as two cells sequenced under the same cellular barcode (e.g., being captured in the same droplet), are fairly frequent in scRNAseq datasets, with estimates ranging from 1 to 10% depending on the platform and cell concentration used [26, 27]. While doublets of the same cell type are relatively innocuous for most downstream applications due to their conservation of the relative expression between genes, doublets formed from different cell types or states are likely to be misclassified and could potentially distort downstream analysis. In some cases, doublets can be identified through their unusually high number of reads and detected features, but this is not always the case (Additional File 1: Figure S4). A number of methods were developed to identify doublets, in most cases by comparing each cell to artificially created doublets [28–30]. We first evaluated the capacity of these methods to detect doublets using the two 10x datasets with cells of different genetic identity [15], using SNP genotypes as the ground truth. For the sole purpose of this section, we included an additional dataset with SNP information but lacking true cell labels [27]. Of note, SNP-based analyses also call doublets created by cells of the same cell type (but from different individuals) and which are generally described as homotypic (as opposed to neotypic or heterotypic doublets, i.e., doublets from different cell types). These homotypic doublets might not be identifiable from the mere gene counts, and their identification is not generally the primary aim of doublet callers since they are often considered innocuous and, when across individuals, can be identified through other means (e.g., SNPs). We therefore do not expect a perfect accuracy in datasets involving cells of the same type across individuals (as in the demuxlet dataset).
We tested DoubletFinder [28] and scran’s doubletCells [29], both of which use similarity to artificial doublets, and scds [30], which relies on a combination of co-expression and binary classification. DoubletFinder integrates a thresholding based on the proportion of expected doublets, while scran and scds return scores that must be manually thresholded. In these cases, we ensured that the right number of cells would be called doublets. In addition to these methods, we reasoned that an approach such as DoubletFinder could be simplified by being applied directly on counts and by using a pre-clustering to create neotypic/heterotypic doublets more efficiently. We therefore also evaluated a simple and fast Bioconductor package implementing this method for doublet detection, scDblFinder, with the added advantage of accounting for uncertainty in the expected doublet rate and using meta-cells from the clusters to even include triplets (see the “Methods” section).
While most methods accurately identified the doublets in the 3 cell lines dataset (mixology10x3cl), the other two datasets proved more difficult (Fig. 2a). scDblFinder achieved comparable or better accuracy than top alternatives while being the fastest method (Fig. 2b). Across datasets, cells called as doublets tended to be classified in the wrong cluster more often than other cells (Fig. 2c). We also found that scDblFinder clearly improved the accuracy of the clustering across datasets that are expected to have heterotypic doublets (mixology datasets) and even in a simulation (simMix1) that should not contain any (Fig. 3). Across the datasets that, being FACS-sorted, should not contain such doublets, doublet removal tended not to robustly impact clustering accuracy. However, in few datasets (ZhengMix4eq and Zhengmix4uneq, and simMix2 with a very small difference), doublet removal performed worse when combined with lenient or no filtering.
Identification of doublet cells. a Receiver operating characteristic (ROC) curves of the tested doublet detection methods for three datasets with SNP-identified doublets. Dots indicate threshold determined by the true number of doublets. b Running time of the different methods (DoubletFinder failed on one of the datasets). c Rate of misclassification of the cells identified by scDblFinder as doublets (DBL) or singlets (SNG), across a large range of clustering analysis. Even in datasets which should not have neotypic doublets, the cells identified as such tend to be misclassified
Effect of filtering on cell subpopulation structure and clustering. a Filtering on the basis of distance to the whole distribution can lead to strong bias against certain subpopulations. The dashed line indicates a threshold of 2.5 median absolute deviations (MADs) from the median of the overall population. b Relationship between the maximum subpopulation exclusion rate and the average clustering accuracy per subpopulation across various filtering strategies.The datasets for which the presence of doublets could be confirmed with SNP genotypes are labeled in bold. Of note, doublet removal appears to have a neutral or positive impact even when, due to the design, there are no heterotypic doublets in the data. The PCA methods refer to multivariate outlier detected as implemented in scater (see the “Methods” section for details)
Excluding more cells is not necessarily better
Beyond doublets, a dataset might include low-quality cells whose elimination would reduce noise. This has for instance been demonstrated for droplets containing a high content of mitochondrial reads, often as a result of cell degradation and resulting loss of cytoplasmic mRNAs [31]. A common practice is to exclude cells that differ considerably from most other cells on the basis of such properties. This can for instance be performed through the isOutlier function from scater that measures, for a given control property, the number of median absolute deviations (MADs) of each cell from the median of all cells. Additional File 1: Figure S5 shows the distributions of some of the typical cell properties commonly used. Of note, these properties tend to be correlated, with some exceptions. For example, while a high proportion of mitochondrial reads is often correlated with a high proportion of the counts in the top features, there can also be other reasons for an over-representation of highly expressed features (Additional File 1: Figure S6), such as an over-amplification in non-UMI datasets. In our experience, 10X datasets also exhibit a very strong correlation between the total counts and the total features even across very different cell types (Additional File 1: Figure S7). We therefore also measure the ratio between the two and treat cells strongly departing from this trend with suspicion.
Reasoning that the cells we wish to exclude are the cells that would be misclassified, we measured the rate of misclassification of each cell in each dataset across a variety of clustering pipelines, correcting for the median misclassification rate of the subpopulation and then evaluated what properties could be predictive of misclassification (Additional File 1: Figure S8–10). We could not identify any property or simple combination thereof that would be consistently predictive of misclassification; the only feature that stood out across more than one dataset (the Zheng datasets) was that cells with very high read counts have a higher chance of being misclassified, although such a pattern was not observed in the other datasets.
We next investigated the impact of several filtering methods (see the “Methods” section for more details): four methods based on deviations to MADs with increasing levels of stringency (named lenient, default, stringent, veryStringent) and two methods based on scater’s runPCA using all (pca.all) or selected covariates (pca.sel). An important risk of excluding cells on the basis of their distance from the whole distribution of cells on some properties (e.g., library size) is that these properties tend to have different distributions across subpopulations. As a result, thresholds in terms of number of MADs from the whole distribution can lead to strong biases against certain subpopulations (Fig. 3a). We therefore examined the tradeoff between the increased accuracy of filtering and the maximum proportion of cells excluded per subpopulation (Fig. 3b and Additional File 1: Figure S11). Since filtering changes the relative abundance of the different subpopulations, global clustering accuracy metrics such as ARI are not appropriate here. We therefore calculated the per-subpopulation precision and recall using the Hungarian algorithm [32] and monitored the mean F1 score across subpopulations. A first observation was that, although more stringent filtering tended to be associated with an increase in accuracy, it tended to plateau and could also become deleterious. Most of the benefits could be achieved without very stringent filtering and minimizing subpopulation bias (Fig. 3b). Applying the same filtering criteria on individual clusters of cells (identified through scran’s quickCluster method) rather than on whole dataset resulted in nearly no cells being filtered out (Additional File 1: Figure S12A). This suggests that stringent filtering on the global population tends to discard cells of subpopulations with more extreme properties (e.g., high library size), rather than low-quality cells. In contrast, too lenient filtering (or lack thereof) can lead to misclassified cells, as was the case for the Zhengmix4 datasets (Fig. 3b). We found that a relatively mild filtering (default—see the “Methods” section) provided a good trade-off, often leading to high clustering performance (e.g., simMix and Zhengmix4 datasets) while retaining most of the cells in each subpopulation (e.g., Koh and Zhengmix8eq). PCA-based selection did not appear superior to feature-wise filters. We found very little overlap between the cells excluded by doublet removal and those excluded by MAD-based filtering (Additional File 1: Figure S12B). Of note, doublet removal in conjunction with filtering tended to improve accuracy not only in datasets that do include heterotypic doublets (mixology datasets), but also some others in which such doublets are unlikely given the experimental design (e.g., FACS-sorted datasets). Indeed, the datasets where doublet removal did not have a clear positive impact (e.g., Koh, Kumar, and the simulations) are those in which heterotypic doublets are not expected. Overall, we therefore recommend the use of doublet removal followed by our default filtering (see the “Methods” section) or a similar approach.
Filtering features by type
Mitochondrial reads have been associated with cell degradation and there is evidence that ribosomal genes can influence the clustering output, hiding other biological structure in the analysis [7]. We therefore investigated whether excluding one category of features or the other, or using only protein-coding genes, had an impact on the ability to distinguish subpopulations (Additional File 1: Figure S13). Removal of ribosomal genes robustly reduced the quality of the clustering, suggesting that they represent real biological differences between subpopulations. Removing mitochondrial genes and restricting to protein-coding genes had a very mild impact.
Normalization and scaling
We next investigated the impact of different normalization strategies. Besides the standard log-normalization included in Seurat, we tested scran’s pooling-based normalization [33], sctransform’s variance-stabilizing transformation [34], normalization based on stable genes [35, 36], and SCnorm [37] (scVI-based normalization [38] was included separately, as it was not meant for this purpose and follows a slightly different flow). In addition to log-normalization, the standard Seurat clustering pipeline performs per-feature unit-variance scaling so that the PCA is not too strongly dominated by highly expressed features. We therefore included versions of the different normalization procedures, with or without a subsequent scaling (sctransform’s variance-stabilizing transformation involves an approach analogous to scaling). Seurat’s scaling function (and sctransform) also includes the option to regress out the effect of certain covariates. We tested its performance by using the proportion of mitochondrial counts and the number of detected features as covariates. Finally, it has been proposed that the use of stable genes, in particular cytosolic ribosomal genes, can be used to normalize scRNAseq [36]. We therefore evaluated a simple linear normalization based on the sum of these genes, as well as nuclear genes.
An important motivation for the development of sctransform was the observation that, even after normalization, the first principal components of various datasets tended to correlate with library size, suggesting an inadequate normalization [34]. However, as library size tends to vary across subpopulations, part of this effect can simply reflect biological differences. We therefore assessed to what extent the first principal component still retained a correlation with the library size and the number of detected features, removing the confounding covariation with the subpopulations (Fig. 4a and Additional File 1: Figure S14). Scaling tended to remove much of the correlation with these features, and while most methods were able to remove most of the effect, the correlation was lowest with sctransform. Surprisingly, however, regressing out the covariates tended to increase the association with it.
Evaluation of normalization procedures. a Variance in PC1 explained by library size (left) and detection rate (right) after accounting for subpopulation differences. b Average silhouette width per true subpopulation, where higher silhouette width means a higher separability. c Clustering accuracy (Seurat clustering), measured by mutual information (MI), Adjusted Rand Index (ARI), and ARI at the true number of cluster. Gray squares indicate that the true number of clusters could not be reached with the corresponding method, dataset, and parameter set. “feat_regress,” “mt_regress,” and “feat_mt_regress” respectively stand for the regressing out of the number of features, the proportion of mitochondrial reads, or both during scaling. Throughout this paper, silhouette plots (b) square root transform values for color mapping. Other evaluation metric heatmaps (e.g., c here) map colors to the signed square root of the number of (matrix-wise) median absolute deviations from the (column-wise) median. Using this mapping, differences in color have the same meaning across datasets, and the color scale is not primarily capturing outliers or baseline differences between datasets. The numbers printed in the cells represent the raw (i.e., unscaled) metric values and are printed in white or black depending on whether they are above or below the median
We further evaluated normalization methods by investigating their impact on the separability of the subpopulations. Since clustering accuracy metrics such as the ARI are very strongly influenced by the number of clusters, we complemented it with silhouette width [39] and mutual information (MI, Fig. 4b, c). We found most methods (including no normalization at all) to perform fairly well in most of the subpopulations and that some datasets (e.g., Zhengmix8eq and simMix1) and subpopulations were consistently more challenging across all methods. An exception was scVI-based normalization, which performed similarly to other methods on some datasets (Kumar, simMix2, Zhengmix4eq, Zhengmix4uneq), but very poorly on others, in terms of both the subpopulation silhouette and clustering accuracy (Additional File 1: Figure S13; of note, these values were not meant to be used for clustering, and the usage of the scVI latent space will be discussed below along with dimensionality reductions). Scaling (e.g., “seurat (no scale)” vs “seurat”) tended to reduce the average silhouette width of some subpopulations and to increase that of some less distinguishable ones and was generally, but not always, beneficial on the accuracy of the final clustering. Regressing out covariates systematically gave equal or poorer performance on MI, ARI, and ARI at true number of clusters, with a strongest negative impact among the Smart-seq datasets (Koh and Kumar). Regressing out covariates using sctransform instead tended to have a much milder effect, possibly owing to its regularization. In general, sctransform systematically outperformed other methods and, even though it was developed to be applied to data with unique molecular identifiers (UMI), it also performed fairly well with the Smart-seq protocol (Koh and Kumar datasets).
Finally, we monitored whether, under the same downstream clustering analysis, different normalization methods tended to lead to an over- or under-estimation of the number of clusters. Although some methods had a tendency to lead to a higher (e.g., sctransform) or lower (e.g., stable genes) number of clusters, the effect was very mild and not entirely systematic (Additional File 1: Figure S16). We also confirmed that, especially in the UMI-based datasets, sctransform did in fact successfully stabilize variance across mean counts (Additional File 1: Figure S17), at little apparent cost on computation time (Additional File 1: Figure S18).
Feature selection
A standard clustering pipeline typically involves a step of highly variable genes selection, which is complicated by the digital nature and the mean-variance relationship of (sc)RNAseq. Seurat’s earlier approaches involved the use of dispersion estimates standardized for the mean expression levels, while more recent versions (≥ 3.0) rely on a different measure of variance, again standardized. While adjusting for the mean-variance relationship removes much of the bias towards highly expressed genes, it is plausible that this relationship may in fact sometimes reflects biological relevance and would be helpful in classifying cell types. Another common practice in feature selection is to use those with the highest mean expression. Recently, it was instead suggested to use deviance [40], while sctransform provides its own ordering of genes based on transformed variance.
Reasoning that a selection method should ideally select genes whose variability is higher between subpopulations than within, we first assessed to what extent each method selected genes with a high proportion of variance or deviance explained by the (real) subpopulations. As the proportion of variability in a gene attributable to subpopulations can be measured in various ways, we first compared three approaches: ANOVA on log-normalized count, ANOVA on sctransform normalization, and deviance explained. The ANOVAs performed on a standard Seurat normalization and on sctransform data were highly correlated (Pearson correlation > 0.9, Additional File 1: Figure S19A). These estimates were also in good agreement with the deviance explained, although lowly expressed genes could have a high deviance explained without having much of their variance explained by subpopulation (Additional File 1: Figure S19B–D). We therefore compared the proportion of the cumulative variance/deviance explained by the top X genes that could be retrieved by each gene ranking method (Additional File 1: Figure S20-21). We first focused on the first 1000 genes to highlight the differences between methods, although a higher number of selected genes decreased the differences between methods (Additional File 1: Figure S20-22). The standardized measures of variability were systematically worse than their non-standardized counterparts in selecting genes with a high proportion of variance explained by subpopulation. Regarding the percentage of deviance explained, however, the standardized measures were often superior (Fig. 5a and Additional File 1: Figure S20-21). Deviance proved the method of choice to prioritize genes with a high variance explained by subpopulations (with mere expression level proving surprisingly performant) but did not perform so well to select genes with a high deviance explained.
Evaluation of feature selection methods. a Ability of different feature ranking methods to capture genes with a high proportion of variance (left) or deviance (right) explained by real subpopulations. The asterisk denotes the default Seurat method. b Accuracy of clusterings (at the true number of clusters) when selecting 1000 genes using the given methods. Based on standard Seurat normalization (left) or sctransform (right). The vst.varExp and devianceExplained methods correspond to the estimates used in a to evaluate the selection methods and were included here only for validation purpose
We next evaluated how the use of different feature selection methods affected the clustering accuracy (Fig. 5b). To validate the previous assay on the proportion of variance/deviance explained by real populations, we included genes that maximized these two latter measures. Interestingly, while these selections were on average the top-ranking methods, they were not systematically best for all datasets (Fig. 5b). Non-standardized measures of variability, including mere expression level, tended to outperform more complex metrics. In general, we found deviance and unstandardized estimates of variance to provide the best results across datasets and normalization methods. Increasing the number of features selected also tended to lead to an increase in the accuracy of the clustering, typically plateauing after 4000 features (Additional File 1: Figure S22).
Dimensionality reduction
Since the various PCA approaches and implementations were recently benchmarked in a similar context [17], we focused on widely used approaches that had not yet been compared: Seurat’s PCA, scran’s denoisePCA, and GLM-PCA [40]. When relevant, we combined them with sctransform normalization. Given that Seurat’s default PCA weights the cell embeddings by the variance of each component, we also evaluated the impact of this weighting with each method.
The impact of the dimensionality reduction methods tested was far greater than that of normalization or feature selection (Additional File 1: Figure S23). Overall, weighting the components by their explained variance tended to increase silhouette width (Additional File 1: Figure S23A) and, in most datasets, clustering accuracy (Additional File 1: Figure S23B). GLM-PCA tended to increase the average silhouette width of already well-defined subpopulations, but Seurat’s PCA procedure however proved superior on all metrics. Like GLM-PCA, scVI’s Linearly Decoded (LD) data [41] and latent space do not explicitly rely on normalized counts. Their performance was however worse than Seurat’s PCA in view of the silhouette width (Additional File 1: Figure S15). In terms of clustering accuracy, scVI’s latent space performed better that its LD but was nevertheless less accurate than PCA-based alternatives on the datasets with a very low amount of cells (Koh and Kumar) and harboring many subpopulations (simMix1 and Zhengmix8eq).
Estimating the number of dimensions
A common step following dimension reduction is the selection of an appropriate number of dimensions to use for downstream analysis. Since Euclidean distance decreases as the number of non-discriminating dimensions increases, there is usually a trade-off between selecting enough dimensions to keep most information and excluding smaller dimensions that may represent technical noise or other unwanted sources of variation. Overall, increasing the number of dimensions robustly led to a decrease in the number of clusters (Additional File 1: Figure S16 and 24). This tended to affect the accuracy of the clustering (Additional File 1: Figure S25), although in both cases (number of clusters and ARI), Seurat’s resolution parameter had a much stronger impact.
Different approaches have been proposed to select the appropriate number of dimensions, from the visual identification of an inflexion point in the curve of the variance explained by each component (“elbow” method), to more complex algorithms. We evaluated the performance of dimensionality estimators implemented in the intrinsicDimension package [42], as well as common procedures such as the “elbow‘’ method, some scRNAseq-specific methods such as the JackStraw procedure [43] or scran’s denoisePCA [29], and the recent application of Fisher Separability analysis [44]. We also included a variation of the molecular cross-validation (MCVR, modified to be applied on normalized data—see the “Methods” section) approach originally developed to select an calibrate denoising methods [45], but which has also been applied to estimate dimensionality [46].
We compared the various estimates in their ability to retrieve the intrinsic number of dimensions in a dataset, based on Seurat’s weighted PCA space. As a first approximation of the true dimensionality, we computed the variance in each principal component (based on Seurat’s weighted PCA space) that was explained by the subpopulations, which sharply decreased after the first few components in most datasets (Fig. 6a). These truth-based estimates were then compared to those of the above dimensionality estimation methods (Fig. 6b). Of note, the methods differ widely in compute time (Fig. 6b) and we saw no relationship between the accuracy of the estimates and the complexity of the method. Reasoning that over-estimating the number of dimensions is less problematic than under-estimating it, we kept the former methods for a full analysis of their impact on clustering (Fig. 6c-d), when combined with sctransform or standard Seurat normalization.
Estimating dimensionality. a Estimated dimensionality using the proportion of variance in each component explained by the true subpopulations. b Difference between “real” dimensionality (from a) and dimensionality estimation methods, along with computing time. c Average silhouette width per subpopulation across a selection of methods, combined with sctransform or standard Seurat normalization. d Clustering accuracy across the normalization/dimensionality estimation methods. The color-mapping scheme is the same as in Fig. 4c
MCVR and the global maximum likelihood based on translated Poisson mixture model (maxLikGlobal) tended to produce clusterings with smaller deviations to the true number of subpopulations (Additional File 1: Figure S26). Although most methods performed well on the various clustering measures, maxLikGlobal (using 10 or 20 nearest neighbors, although estimates were relatively stable across various values of k—see Additional File 1: Figure S27) provided the dimensionality estimate that best separated challenging subpopulations (Fig. 6c) and tended to result in the best clustering accuracy (Fig. 6d). MCVR did not perform well for the datasets without UMIs (Koh and Kumar), which violates the assumptions of the method, and while it did increase the separability of some subpopulations (Fig. 6c), it was detrimental to the classification of some others. We observed a strong negative correlation (r = − 0.995, p 1.6e −8) between the number of dimensions called by MCVR and the log10 average library size of a dataset, which explained the former considerably better than the number of subpopulations.
Of note, the optimal method in our evaluation (maxLikGlobal) systematically selected many more components than were associated with the subpopulations in Fig. 6a, suggesting that although these additional components appear individually uninformative, in combination, they nevertheless contribute to classification. An important implication is that the commonly used elbow method most likely underestimates dimensionality.
Clustering
The last step evaluated in our pipeline was clustering. Given previous work on the topic [6, 7] and the success of graph-based clustering methods for scRNAseq, we restricted our evaluation to Seurat’s method and variations of scran’s graph-based clustering using random walks (walktrap method) or the optimization of the modularity score (fast_greedy) on various underlying graphs. Specifically, we compared rank-based graphs (scran’s default) with graphs using the components’ values directly, SNN vs KNN graphs, and the exact nearest neighbors to the Annoy approximation. Again, the tested methods were combined with Seurat’s standard normalization and sctransform, otherwise using the parameters found optimal in the previous steps.
Since ARI is dominated by differences in the number of clusters (Additional File 1: Figure S2–3) and no single metric is perfect, we diversified them (Fig. 7). MI has the virtue of not decreasing when a true subpopulation is split into two clusters, which is arguably less problematic (and might well reflect unknown biological subgroups), but as a consequence, it can be biased towards methods producing higher resolution clustering (Additional File 1: Figure S3). Similarly, precision per true subpopulation is considerably more robust to differences in the number of clusters, and we also tracked the ARI at the true number of clusters.
Evaluation of clustering methods. Clustering accuracy (a) and run time (b) of scran- and Seurat-based clustering tools in combination with sctransform and standard Seurat’s normalization. The color-mapping scheme in a and the left of b is the same as in Fig. 4c
The MI score, which is largely independent of the estimated number of clusters, was overall higher for the walktrap method (Fig. 7a, left) over Seurat and the fast-greedy method. Depending on what network it was used with, however, it sometimes yielded a lower ARI or minimum precision. The ARI score at the true number of clusters, when available, showed similar performances, especially when using sctransform Because Seurat’s resolution parameter had a large impact on the number of clusters identified (Additional File 1: Figure S2 and 24), Seurat could always be coerced into producing the right number of clusters. Instead, the number of clusters found by scran was considerably less influenced by the available parameters (number of nearest neighbors or steps in the random walk—see Additional File 1: Figure S28), and as a result, scran-based clustering sometimes never produced a partitioning with the right number of clusters. This observation, along with scran’s higher MI score, suggest that scran sometimes simply divides a real subpopulation into two clusters (possibly tracking some unknown biological differences) rather than committing misclassification errors. Overall, the walktrap method appeared superior to the fast greedy algorithm and was generally comparable to Seurat clustering, although the latter offered more control over the resolution. A major difference between walktrap-based clustering and Seurat is the computing time (Fig. 7b). We found that using the Annoy approximation to the nearest neighbors somewhat reduced computing time at no apparent cost to accuracy. It nevertheless remained considerably slower than Seurat. For all methods, some poorly distinguishable subpopulations from both the Zhengmix8eq and simMix1 datasets remained very inaccurately classified in regard to all metrics.
Overview of interactions between steps
We selected the most important alternatives for each parameter (see the “Methods” section) in order to explore their interactions. Unsupervised clustering on the evaluation metrics reveals general patterns in the results as well as the relative importance of the parameters (Additional File 1: Figure S29), with the clustering method having the most important impact on the results. This is however chiefly due to number of clusters called: an ANOVA on all analyses after accounting, in the model, for baseline differences between datasets and the impact of Seurat’s resolution parameter, attributed relatively little importance to the clustering method, and instead attributed the largest proportion of variance in the main cluster metrics to the normalization method and the number of dimensions used (Fig. 8). Part of the impact of the estimated dimensionality depended on its effect on the number of clusters detected, and it indeed explained less variance in ARI at the true number of clusters (Fig. 8). Nevertheless, the choice of dimensionality (as well as feature selection) came closely behind normalization, suggesting that more attention should be paid to this relatively neglected step.
Variance in clustering metrics explained by the parameters. Proportion of variance explained by the parameters of the main analysis steps. The proportions are calculated from an ANOVA using the model metric ∼ dataset*resolution+doubletmethod+filt+norm+clustmethod+dims and substracting the variance attributable to differences due to datasets, resolutions, and their interactions
The greater importance of normalization, dimensionality, and feature selection over filtering steps is also visible in the clustering (Additional File 1: Figure S29). The top methods (bottom of Additional File 1: Figure S29) confirm the results of the previous sections, highlighting especially sctransform, deviance-based feature selection, and maxLikGlobal dimensionality estimates as top methods. Interestingly, while the combination of sctransform with Seurat clustering was particularly performant for most datasets, for the simulated “simMix1” dataset, standard Seurat normalization combined with scran-based clustering proved superior.
We next investigated whether the benefits of some methods were conditional on the choices at other steps. Although analysis of variance identified some significant interactions (Additional File 1: Figure S30), we found that the benefits brought by performant methods were robust to differences in other parts of the pipeline (e.g., Additional File 1: Figure S31). For instance, we confirmed that doublet removal and sctransform normalization were beneficial independently of other steps and that deviance-based feature selection outcompeted Seurat’s default selection method independently of changes at other steps (Additional File 1: Figure S31). ANOVA did reveal some significant (negative) interactions (Additional File 1: Figure S30): the top two-way interaction was between scran normalization and the elbow method for selection the number of dimensions, and indeed that combination was involved in all of the 30 lowest-ranking pipelines. Furthermore, all significant interactions involved the elbow method, suggesting that some methods were less robust to underestimated dimensionality.
Further extensions to the pipeline: imputation/denoising
The basic pipeline presented here can be extended with additional analysis steps while keeping the same evaluation metrics. To demonstrate this, we evaluated various imputation or denoising techniques based on their impact on classification. Since preliminary analysis showed that all methods performed equally well or better on normalized data, we applied them after filtering and normalization, but before scaling and reduction. Although some of the methods (e.g., DRImpute_process and alra_norm) did improve the separability of some more elusive subpopulations, no method had a systematically positive impact on the average silhouette width across all subpopulations (Additional File 1: Figure S32A). When restricting ourselves to clustering analyses that yielded the “right” number of clusters, all tested methods improved classification compared to a scenario with no imputation step (“none” label, Additional File 1: Figure S32B). However, the situation was not so straightforward with alternative metrics, where some methods (e.g., ENHANCE) consistently underperformed. 10X datasets, which are typically characterized by a lower per-cell coverage and feature detection rate, benefited more from imputation and, in this context, DrImpute and DCA tended to show the best performance. On the contrary, imputing on normalized counts originating from Smart-seq technology was instead rather deleterious to the clustering accuracy.
Application of pipeComp to a different context
To illustrate the applicability of pipeComp to a completely different problem, we benchmarked the impact of surrogate variable analysis (SVA)-based methods on bulk RNAseq differential expression analysis (DEA). The rationale behind such approaches is that the identification of factors determining genome-wide variation (i.e., surrogate variables) and their inclusion in the DEA’s generalized linear model might improve DEA, especially in the presence of sources of variation unrelated to the experimental variables. sva [47, 48] and RUVseq [49] are two popular packages each implementing different methods to infer these variables. We therefore created a PipelineDefinition with three steps: (1) feature filtering, (2) removal of unwanted variation (i.e., SVA), and (3) DEA using the most established methods [50]. The package’s “pipeComp_dea” vignette details the creation process. We developed 3 benchmark datasets: (1) the ipsc data is a random selection of 10 vs 10 samples from [51], which are very heterogeneous, and foldchanges were manually added to 300 genes; (2) the seqc dataset contains 5 vs 5 samples of two mixtures of the SEQC consortium with different ERCC spike-in mixes [52], and includes a partially correlated batch effect; (3) a simulation of 8 vs 8 samples with two technical vectors of variation (see the “Methods” section for more details). As previously reported [52], we found that more stringent filtering improved DEA (Additional File 1: Figure S33A–B) and that the different DEA methods had similar performances, especially after using SVA-based methods (Additional File 1: Figure S33B). Including a SVA-based step increased sensitivity across all datasets (Fig. 9a and Additional File 1: Figure S33B), particularly in datasets with technical vectors of variation (seqc and simulation), and benefited all DEA methods in a similar fashion (Fig. 9c and Additional File 1: Figure S33C–34A). With the exception of the RUVr method, which led to a high false discovery rate (FDR), all other methods showed comparable gains in sensitivity while maintaining a decent error control (if a little above the nominal FDR), even when increasing the number of surrogate variables used (Fig. 9b–c). While the FDR did increase in the ipsc dataset, we suspect that much of the effect could be due to spurious differences [53] between the groups which we wrongly assume to be false positives. Nevertheless, the FDR remained acceptable with up to three variables. These results therefore suggest that these methods are robust and can be widely applied.
Evaluation of the impact of surrogate variable analysis (SVA) on (bulk) RNAseq differential expression analysis using pipeComp. RUVr and RUVs refer to the two RUVSeq functions by the same name; svaseq refers to the function of the same name in sva, while vstsva refers to the original sva method applied no variance-stabilized data. a True positive rates (TPR) and false discovery rates (FDR) of the SVA-based methods (using a single surrogate variable), averaged across DEA methods. b TPR and FDR of the different SVA-based methods (averaged across the different DEA methods) using increasing numbers of variables. In a, b, the x-axis is square root transformed to improve readability, and three dots for each combination of methods refer to the nominal FDR thresholds of 0.01, 0.05, and 0.1. c Correlation of estimated logFC with expected ones, as well as TPR and FDR (at nominal FDR threshold of 0.05) of the different methods with increasing numbers of variables. As in Fig. 4c, the color mapping tracks the number of (matrix-wise) median absolute deviations from the (column-wise) median, while the printed numbers represent the raw metric values. be and leek refer to the two methods implemented in the sva package for estimating the number of surrogate variables
Discussion
Practical recommendations
On the basis of our findings, we can make a number of practical analysis recommendations, also summarized in Additional File 1: Figure S35:
-
1
Filtering
-
Doublet detection and removal is advised and can be performed at little computing cost with software such as scDblFinder or scds.
-
Distribution-based cell filtering fails to capture doublets and should use relatively lenient cutoffs (e.g., 5 MADs, or 3 MADs in at least 2 distributions—see for instance our default filters in the the “Methods” section) to exclude poor-quality cells while avoiding bias against some subpopulations.
-
Features filtering based on feature type did not appear beneficial.
-
-
2
Normalization and scaling
-
Most normalization methods tested yielded a fair performance, especially when combined with scaling, which tended to have a positive impact on clustering.
-
sctransform offered the best overall performance in terms of the separability of the subpopulations, as well as removing the effect of library size and detection rate.
-
The common practice of regressing out cell covariates, such as the detection rate or proportion of mitochondrial reads nearly always had a negative impact, leading to increased correlation with covariates and decreased clustering accuracy. We therefore advise against this practice.
-
-
3
Feature selection
-
Deviance [40] offered the best ranking of genes for feature selection.
-
Increasing the number of features included tended to lead to better classifications, plateauing from 4000 features in our datasets.
-
-
4
Denoising/imputation
-
Denoising appeared beneficial to the identification of some subpopulations in 10x datasets, but not in Smart-seq datasets.
-
We found especially ALRA (with prior normalization), DrImpute (with prior processing), and DCA to offer the best performances, although none of these methods was beneficial over all subpopulations.
-
-
5
PCA
-
Similarly to previous reports [14], we recommend the irlba-based PCA using weighting of the components, as implemented in Seurat.
-
We advise against the commonly used elbow method (because it is too conservative) and the jackstraw method (low performance at great computational costs) for deciding on the components to include and recommend a method like the global maximum likelihood based on translated Poisson mixture model method (e.g., implemented in the maxLikGlobalDimEst function of intrinsicDimension, in our case using the 10 or 20 nearest neighbors).
-
-
6
Clustering
-
In cases where prior knowledge can guide the choice of a resolution, Seurat can be useful in affording manual control of it while, in the absence of such knowledge, scran-based walktrap clustering provided reasonable estimates, although at greater computational costs.
-
Limitations and open questions
In this study, we evaluated tools commonly used for the processing of scRNAseq with a focus on droplet-based datasets, namely from the 10x technology. Although this platform has been used in almost half of the scRNAseq studies in 2019 [2], other popular technologies such as Drop-seq, InDrops, or Smart-seq2/3 were not represented in the present benchmarking. Differences between such protocols, even among droplet-based technologies, can have a very large impact on cell capture efficiency, cell numbers, and clustering [54–56]. Although most top-ranking methods in our comparison performed well on both Smart-seq and 10x datasets that we tested, future benchmarking efforts should strive to include less represented technologies. In addition, we did not compare any of the alignment and/or quantification methods used to obtain the count matrix, which was for instance discussed in [18]. Some steps, such as the implementation of the PCA, were also not explored in detail here as they have already been the object of recent and thorough study elsewhere [14, 17]. We also considered only methods relying on Euclidean distance, while correlation was recently reported to be superior [57] and would require further investigation.
Among all tested pipelines, performance varied greatly across datasets. The ZhengMix8eq and simMix1 datasets were the most challenging datasets across all preprocessing steps that we tested. The average silhouette width of half of their subpopulations was systematically lower than the other subpopulations/datasets and the ARI score at true number of clusters, if ever achieved, was always poor. It is likely that these two datasets are challenging due to their combined sparsity and complexity (see Additional File 1: Figure S1). The challenge of distinguishing subpopulations of T-cells in the Zhengmix8eq dataset was previously discussed [6]. In contrast, while the Koh dataset harbors as many populations, they originate from more distinct groups (from hESC in different differentiation stages), while the Kumar and mixology datasets harbor a very low subpopulation complexity and most methods applied on them performed well.
Our recommendations are in line with the results of several other benchmarking studies. For instance, Seurat and scran methods were already shown to yield good clustering performance [7] and DrImpute to improve the quality of downstream analyses [21]. A previous systematic comparison [18] instead found that the use of SAVERX for denoising data and scran for clustering yielded better performance on the ability to recover differentially expressed genes, whereas we focused on the ability to recover cell populations. Another disagreement with the present recommendations come from the study of Hou et al. [10] that recommends SAVERX and discourage the use of DCA for better clustering, whereas our study found opposite results for these tools. Several differences in the design of our study could explain this contrast, such as the use of different metrics (ARI at true number of clusters, mean precision/ recall), of different datasets (Koh, Kumar, simMix simulations) and the use of different filtering and normalization strategies. ALRA however yielded good results in both studies and may be the most conservative option. The study from Hou et al. also highlighted the usefulness of ALRA when followed by differential and trajectory analysis both in UMI and Fluidigm technologies.
The performance of scVI in our evaluation was lower than in the original publication presenting the method [38]. Our results are however in line with other studies comparing silhouette score [58] and clustering accuracy [10] of scVI’s latent space. These studies, like ours, used datasets with relatively few cells (i.e., fewer than genes), for which scVI was reported by its authors to be less adapted. We think that further work will be required to assess its performance on larger datasets.
Here, we chose to concentrate on what could be learned from datasets with known cell labels (as opposed to labels inferred from the data, as in [54]). In contrast to Tian et al. [15], who used RNA mixtures of known proportions, we chose to rely chiefly on real cells and their representative form of variability. Given the limited availability of such well-described datasets, however, several aspects of single-cell analysis could not be compared, such as batch effect correction or multi-dataset integration. For these aspects of scRNAseq processing that are critical in some experimental designs, we refer the reader to previous evaluations [16, 59]. In addition, a focus on the identification of the subpopulations might fail to reveal methods that are instead best performing for tasks other than clustering, such as differential expression analysis or trajectory inference. Several informative benchmarks have already been performed on some of these topics [5, 11, 13, 19, 22, 60]. Yet, such evaluations could benefit from considering methods not in isolation, but as parts of a connected workflow, as we have done here. We believe that the pipeComp framework is modular and flexible enough to integrate new steps in the pipeline, as shown by an example with imputation/denoising methods.
We developed pipeComp to address the need of a framework that can simultaneously evaluate the interaction of multiple tools. The work of Vieth and colleagues[18] already offered an important precedent in this respect, evaluating the interaction of various steps in the context of scRNAseq differential expression analysis, but did not offer a platform for doing so. Instead, the CellBench package was recently proposed to address a similar need [61], offering an elegant piping syntax to combine alternative methods at successive steps. A key additional feature of pipeComp is its ability to perform evaluation in parallel to the pipeline and offering on-the-fly evaluation at multiple steps. This avoids the need to store potentially large intermediate data for all possible permutations and thus allowing a combinatorial benchmark. This, along with the set-up simplicity (demonstrated here by our separate SVA-DEA benchmark), also distinguishes pipeComp from general workflow frameworks such as drake [62]; indeed, we estimate that the benchmarks discussed here would have taken approximately 16TB of storage had all non-redundant intermediates been saved (see the Additional File 2 for more detailed explanations of the framework and comparison to CellBench and drake). In addition, the fact that benchmark functions are stored in the PipelineDefinition makes the benchmark more smoothly portable, making it easy to modify, extend with further methods, or apply to other datasets.
With respect to the methods themselves, we believe there is still space for improvement in some of the steps. Concerning cell filtering, we noticed that the current approach based on whole-population characteristic (e.g., MAD cut-off) can be biased against certain subpopulations, suggesting that more refined methods expecting multimodal distributions should be used rather than relying on whole-population characteristics. In addition, most common filtering approaches do not harness the relationship between cell QC properties. Finally, imputation had a varied impact on the clustering analysis and seemed to be linked to the technology that was used to generate the data. Also, the good performance of DrImpute is in line with a previous study focused on the preservation of data structure in trajectory inference from scRNAseq [21] (ALRA was however not included in this previous benchmark). Our results also align with the hypothesis of this study on the respective strengths of linear and non-linear models and their use to different types of data, such as the highest performance of non-linear methods in developmental studies.
Conclusions
pipeComp is a flexible R framework for evaluating methods and parameter combinations in a complex pipeline, computing on-the-fly multilevel evaluation metrics. Applying this framework to scRNAseq clustering enabled us to make clear recommendations on the steps of filtering, normalization, feature selection, denoising, dimensionality reduction, and clustering (Additional File 1: Figure S35). We demonstrate how a diversity of multilevel metrics can be more robust, more sensitive, and more nuanced than simply evaluating the final clustering performance. We further showed that the pipeComp framework can be applied to extend the current field of benchmarking, as well as to investigate pipelines other domains with multi-step computational analyses.
Methods
Simulated datasets
The simMix1 dataset was based on the human PBMC CITE-seq data deposited under accession code GSE100866 of the Gene Expression Omnibus (GEO) website. Both RNA and ADT count data were downloaded from GEO and processed independently using Seurat. We then considered cells that were in the same cluster both in the RNA-based and ADT-based analyses to be real subpopulations and focused on the 4 most abundant ones. We then performed 3 sampling-based simulations with various degrees of separation using muscat [22] and merged the three simulations. The simMix2 dataset was generated from the mouse brain data published in [22] in a similar fashion. The specific code is available on https://github.com/markrobinsonuzh/scRNA_pipelines_paper.
Default pipeline parameters
Where unspecified, the following default parameters or parameter sets were used:
-
scDblFinder was used for doublet identification
-
The default filter sets (see below) were applied
-
Standard Seurat normalization was employed
-
Seurat (≥3.0) variable feature selection was employed, selecting 2000 genes
-
Standard Seurat scaling and PCA were employed
-
To study the impact of upstream steps on clustering, various Seurat clustering analyses were performed, selecting different numbers of dimensions (5, 10, 15, 20, 30, and 50) and using various resolution parameters (0.005, 0.01, 0.02, 0.05, 0.1, 0.15, 0.2, 0.3, 0.4, 0.5, 0.8, 1, 1.2, 1.5, 2, 4). The range of resolution parameters was selected to ensure that the right number of clusters could be obtained in all datasets.
Denoising/imputation
Most imputation/denoising methods were run with the default parameters with the following exceptions; DrImpute documentation advises to process the data prior to imputation by removing lowly expressed genes and cells expressing less than 2 genes. As it is not clear if this step is a hard requirement for the method to perform well, DrImpute was run with and without prior processing (DrImpute_process and DrImpute_noprocess labels, respectively). The DCA method only accepts integers counts, while two of the datasets had non-integer quantification of expected counts. For these datasets, we rounded up the counts prior to imputation. ALRA is designed for normalized data but as we are evaluating normalization downstream to imputation, we used the method on both non-normalized (alra label) and normalized counts (alra_norm label). scImpute requires an estimation of the expected number of clusters with the input data. As the estimation of the true number of cluster may not be known by the user, we evaluated the tool using the true number of clusters (scImpute label) and using an over/under-estimation of this number (scImpute_plus5 and scImpute_min5 labels, respectively). ENHANCE uses a k-nearest neighbor aggregation method and automatically estimates the number of neighbors to merge prior to the imputation. With the smallest datasets, this parameters was estimated to be 1, which lead to an early stop of the function. In such cases, we manually set this parameter to 2 for the method to work.
Dimensionality estimates
For the methods that produce local dimensionality estimates, we used the maximum. For the elbow method, we implemented an automatic procedure by taking the farthest point from a line drawn between the variance explained by the first and last (i.e., 50th) components calculated. For the JackStraw method, since the Seurat documentation advises not to use a p value threshold (which would typically yield a very large number of dimensions) but rather look for a drop in significance, we applied the same farthest point algorithm on the log10(p values), which in our hands reproduced manual threshold selection.
For the molecular cross-validation approach, we relied on a R implementation by Matthew Young (https://github.com/constantAmateur/MCVR), with minor modifications (see wrapper in the pipeComp package). Briefly, since there is no a priori relationship between the dimensionality of the count data and that of the PCA based on the normalized data, MCVR does not use the original approach based Poisson loss, but the mean squared error of the normalized data, and uses balanced 50-50 splits. Because of the 50% split, dimensionality may be underestimated, and titration did suggest that was the case for some of our datasets.
Filter sets
The default set of filters excludes cells that are outliers according to at least two of the following thresholds: log10_total_counts >2.5 MADs or <5 MADs, log10_total_features >2.5 MADs or <5 MADs, pct_counts_in_top_20_features > or <5 MADs, featcount_dist (distance to expected ratio of log10 counts and features) > or <5 MADs, pct_counts_Mt >2.5 MADs and >0.08.
The stringent set of filters uses the same thresholds, but excludes a cell if it is an outlier on any single distribution. The veryStringent set of filters excludes any cell that is an outlier by at least 2 MADs (lower or higher) on any of the following: log10_total_counts, log10_total_features, pct_counts_Mt, or pct_counts_in_top_20_features. The lenient set of filters excludes cells that are outliers on at least two distributions by at least 5 MADs, except for pct_counts_Mt where the threshold is >3 MADs and >0.08.
For cluster-wise filters, clusters were first identified with scran::quickCluster and the filters were then applied separately for each cluster.
The “pca.all” and “pca.sel” clusters refer to the multivariate outlier detection methods implemented in scater, running runPCA with use_coldata=TRUE, detect_outliers=TRUE. “pca.all” uses all covariates, while “pca.sel” uses only the log10(counts), log10(features), proportion mitochondrial, and proportion in the top 50 features.
Variance and deviance explained
Unless specified otherwise, the variance in gene expression explained by subpopulations was calculated on the data normalized and transformed through sctransform. For each gene, we fitted a linear model using the subpopulation as only independent variable (∼subpopulation) and used the R-squared as a measure of the variance explained. The same method was used for principal components.
The deviance explained by subpopulations was calculated directly on counts using the getDevianceExplained function of pipeComp. The function uses edgeR to fit two models, a full (∼librarySize+population) and a reduced one (∼librarySize). For each gene, the deviance explained is then the difference between the deviance of each models, divided by the deviance of the reduced model. In the rare cases where this resulted in a negative deviance explained, it was set to zero.
To estimate the correlation between the principal components and covariates such as library size, we first fitted a linear model on the subpopulations and correlated the residuals of this model with the covariate of interest.
scVI normalization and LD
The evaluation of scVI algorithm was performed separate to the other pipeline combinations since the model training on our reference datasets was especially resource-consuming and follows a different workflow from the other pipelines. In particular, for datasets with fewer cells than genes, scVI requires a feature selection to be performed prior to the normalization step; furthermore, its dimensional reduction steps rely directly on raw counts. We therefore evaluated it separately and compared it to a representative subset of pipelines using sctransform normalization and Seurat’s PCA dimension reduction method. The resulting evaluation is available in the Additional File 1: Figure S13.
Using scVI, we considered as normalized values the normalized mean of the negative binomial distribution modeling the counts (accessed through the get_sample_scale method). Following scVI’s online documentation, we used the VAE model, using a train size of 1, a training on 400 epochs, and a learning rate of 0.001. As a major limitation of scVI is the training on dataset with low gene to cell ratio, the features were subsampled to 2000 using the subsample_genes function, which resembles the Seurat V3 VST gene selection method. All other parameters were left unmodified.
Two models of the scVI package computing a dimension reduction on the data were tested. First, we evaluated the mean of the posterior distribution for the latent space, still based on the VAE model (accessed through the get_latent function). All other parameters were set as described above or left as default. The second model that we evaluated was the linearly decoded VAE of scVI (LDVAE model). It was trained on 250 epochs, a learning rate of 0.001 and computed 50 dimensions. For some smaller datasets, the training phase aborted. In such cases, we followed the recommendations of the package and reduced the learning rate by a factor 10 until the model could properly converge. Given that the other methods that we evaluated returned PCs ordered by their variance explained, we applied to the same reordering to the latent space returned by scVI. For both models. we relied on the feature selections results from the pipeline and not on the subsample_genes function. All other parameters were left unmodified.
Overall run for interactions across steps
The set of alternatives found for the overall run (Fig. 8 and Additional File 1: Figure S29-31) can be found in the paper’srepository . The analysis of variance was performed with the following model:
metric ∼ dataset*resolution+doubletmethod+filt+norm+ clustmethod+dims
SVA-DEA benchmark
The full process of creating the SVA-DEA PipelineDefinition is described step by step in the pipeComp_deavignette .
The three benchmark datasets are available in https://github.com/markrobinsonuzh/scRNA_pipelines_paper/blob/master/svadea/datasets/, along with all code used to generate and prepare them. Briefly:
-
For the ipsc dataset, we took the quantification of the GSE79636 dataset (fairly heterogeneous) from the iPSCpower package [53] and selected two random sets of 10 samples. No further variation was added, and log-foldchanges ranging from 0.25 to 1.25 were added to the counts of 300 random genes. All other genes were assumed to be negatives.
-
For the seqc dataset, we used data from the seqc package [52]. For both groups (mixtures C and D, which also contain two different spike-in mixes), we selected samples from the ILM_refseq_gene_AGR and ILM_refseq_gene_CNL batches, which have clear technical differences; we selected 5 samples per group with a weak correlation with the batch (3:2 vs 2:3). Since the true differences between mixtures are not entirely known, the analysis was performed on all genes but only the spike-ins were considered for benchmarking.
-
For the simulation, we simulated counts using the polyester package [63] and the means/dispersions from the GSE79636 dataset (restricted to a single batch and biological group), with 500 DEGs (log-foldchanges ranging from 0.5 to 2) and two batch effects: (1) a technical batch partially correlated with the group (6:2) and affecting 1/3 of the genes and (2) a linear vector of variation uncorrelated with the groups and affecting 1/3 of the genes.
Filtering was performed with edgeR’s filterByExpr function, changing the min.count argument. For vstsva, we first applied the variance-stabilizing transformation from DESeq2 [64] before applying sva::sva. edgeR.QLF refers to the quasi-likelihood version [65] of edgeR’s fit. The exact wrappers used can be found in the package at https://github.com/plger/pipeComp/blob/master/inst/extdata/dea_wrappers.R.
Software and package versions
Analyses were performed in R 3.6.0 (Bioconductor 3.9) and the following packages were installed from GitHub repositories: Seurat (version 3.0.0), sctransform (0.2.0), DoubletFinder (2.0.1), scDblFinder (1.1.4), scds (1.0.0), SAVERX (1.0.0), scImpute (0.0.9), ALRA (commit 7636de8), DCA (0.2.2), DrImpute (1.2), ENHANCE (R version, commit 1571696), and SAVERX (1.0.0). The glmPCA was performed using the glmpca package, while the code for deviance calculation was obtained from https://github.com/willtownes/scrna2019 (commit 1ddcc30ebb95d083a685f12fe81d35dd1b0cb1b2).
Availability of data and materials
All analyses were performed through the pipeComp R package, which implements the pipeline framework described here. scVI results and results of the clustering section were produced with pipeComp 0.99.4, while all other results were produced with version 0.99.2 (biorxiv) (branch of the repository). All code to reproduce the simulations and figures is available in the https://github.com/markrobinsonuzh/scRNA_pipelines_paper repository, which also includes the basic datasets with a standardized annotation. All wrappers for methods used in the study can be found in the external data of the pipeComp package (https://github.com/plger/pipeComp/tree/master/inst/extdata). In addition, both the package and attending code are archived on [66].
The gene counts for the two mixology datasets were downloaded from the CellBenchrepository, commit 74fe79e. For the other datasets, we used the unfiltered counts from [6], available on the corresponding repository https://github.com/markrobinsonuzh/scRNAseq_clustering_comparison. For simplicity, all starting SingleCellExperiment objects with standardized metadata are available on [67].
All data and code are released under a GNU General Public License.
References
Zappia L, Phipson B, Oshlack A. Exploring the single-cell RNA-seq analysis landscape with the scRNA-tools database. PLoS Comput Biol. 2018; 14(6):1006245. https://doi.org/10.1371/journal.pcbi.1006245.
Svensson V, Beltrame E. d. V., Pachter L. A curated database reveals trends in single cell transcriptomics. bioRxiv. 2019:742304. https://doi.org/10.1101/742304.
Cobos FA, Alquicira-Hernandez J, Powell J, Mestdagh P, De Preter K. Comprehensive benchmarking of computational deconvolution of transcriptomics data. bioRxiv. 2020. https://doi.org/10.1101/2020.01.10.897116.T.
Cole MB, Risso D, Wagner A, DeTomaso D, Ngai J, Purdom E, Dudoit S, Yosef N. Performance assessment and selection of normalization procedures for single-cell RNA-Seq. Cell Syst. 2019; 8(4):315–28. https://doi.org/10.1016/j.cels.2019.03.010.
Dal Molin A, Baruzzo G, Di Camillo B. Single-cell RNA-sequencing: Assessment of differential expression analysis methods. Front Genet. 2017; 8(62). https://doi.org/10.3389/fgene.2017.00062.
Duo A, Robinson MD, Soneson C. A systematic performance evaluation of clustering methods for single-cell RNA-seq data. F1000Research. 2018; 7:1141. https://doi.org/10.12688/f1000research.15666.2.
Freytag S, Tian L, Lönnstedt I, Ng M, Bahlo M. Comparison of clustering tools in R for medium-sized 10x Genomics single-cell RNA-sequencing data. F1000Research. 2018; 7(1297):1–29. https://doi.org/10.12688/f1000research.15809.2.
Gao M, Ling M, Tang X, Wang S, Xiao X, Qiao Y, Yang W, Yu R. Comparison of high-throughput single-cell RNA sequencing data processing pipelines. bioRxiv. 2020. https://doi.org/10.1101/2020.02.09.940221.
Heiser CN, Lau KS. A quantitative framework for evaluating single-cell data structure preservation by dimensionality reduction techniques. bioRxiv. 2019:684340. https://doi.org/10.1101/684340.
Hou W, Ji Z, Ji H, Hicks SC. A systematic evaluation of single-cell RNA-sequencing imputation methods. bioRxiv. 2020. https://doi.org/10.1101/2020.01.29.925974.
Jaakkola MK, Seyednasrollah F, Mehmood A, Elo LL. Comparison of methods to detect differentially expressed genes between single-cell populations. Brief Bioinforma. 2017; 18(5):735–43. https://doi.org/10.1093/bib/bbw057.
Krzak M, Raykov Y, Boukouvalas A, Cutillo L, Angelini C. Benchmark and parameter sensitivity analysis of single-cell RNA sequencing clustering methods. Front Genet. 2019; 10:1253. https://doi.org/10.3389/fgene.2019.01253.
Soneson C, Robinson MD. Bias, robustness and scalability in single-cell differential expression analysis. Nat Methods. 2018; 15(4):255–61. https://doi.org/10.1038/nmeth.4612.
Sun S, Zhu J, Ma Y, Zhou X. Accuracy, robustness and scalability of dimensionality reduction methods for single-cell RNA-seq analysis. Genome Biol. 2019; 20(269):1–21. https://doi.org/10.1186/s13059-019-1898-6.
Tian L, Dong X, Freytag S, Le Cao K-A, Su S, Amann-Zalcenstein D, Weber TS, Seidi A, Naik S, Ritchie ME. scRNA-seq mixology: towards better benchmarking of single cell RNA-seq protocols and analysis methods. bioRxiv. 2018:433102. https://doi.org/10.1101/433102.
Tran HTN, Ang KS, Chevrier M, Zhang X, Lee NYS, Goh M, Chen J. A benchmark of batch-effect correction methods for single-cell RNA sequencing data,. Genome Biol. 2020; 21(1):1–32. https://doi.org/10.1186/s13059-019-1850-9.
Tsuyuzaki K, Sato H, Sato K, Nikaido I. Benchmarking principal component analysis for large-scale single-cell RNA-sequencing. Genome Biol. 2020; 21(9):1–17. https://doi.org/10.1186/s13059-019-1900-3.
Vieth B, Parekh S, Ziegenhain C, Enard W, Hellmann I. A systematic evaluation of single cell RNA-seq analysis pipelines. Nat Commun. 2019; 10(1):1–11. https://doi.org/10.1038/s41467-019-12266-7.
Wang T, Li B, Nelson CE, Nabavi S. Comparative analysis of differential gene expression analysis tools for single-cell RNA sequencing data. BMC Bioinformatics. 2019; 20.1(40):1–16. https://doi.org/10.1186/s12859-019-2599-6.
Yip SH, Sham PC, Wang J. Evaluation of tools for highly variable gene discovery from single-cell RNA-seq data. Brief Bioinforma. 2018; 20(4):1583–9. https://doi.org/10.1093/bib/bby011.
Zhang L, Zhang S. Comparison of computational methods for imputing single-cell RNA-sequencing data. IEEE/ACM Trans Comput Biol Bioinforma. 2018. https://doi.org/10.1109/TCBB.2018.2848633.
Crowell HL, Soneson C, Germain P-L, Calini D, Collin L, Raposo C, Malhotra D, Robinson MD. On the discovery of population-specific state transitions from multi-sample multi-condition single-cell RNA sequencing data. bioRxiv. 2019;:713412. https://doi.org/10.1101/713412.
Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015; 33(5):495–502. https://doi.org/10.1038/nbt.3192.
Hubert L, Arabie P. Comparing partitions. J Classif. 1985; 2(1):193–218. https://doi.org/10.1007/BF01908075.
Steinley D. Properties of the hubert-arable adjusted rand index. Psychol Methods. 2004; 9(3):386–96. https://doi.org/10.1037/1082-989X.9.3.386.
Bloom JD. Estimating the frequency of multiplets in single-cell RNA sequencing from cell-mixing experiments. PeerJ. 2018; 6:5578. https://doi.org/10.7717/peerj.5578.
Kang HM, Subramaniam M, Targ S, Nguyen M, Maliskova L, McCarthy E, Wan E, Wong S, Byrnes L, Lanata CM, Gate RE, Mostafavi S, Marson A, Zaitlen N, Criswell LA, Ye CJ. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat Biotechnol. 2018; 36(1):89–94. https://doi.org/10.1038/nbt.4042.
McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019; 8(4):329–37. https://doi.org/10.1016/j.cels.2019.03.003.
Lun ATL, Mccarthy DJ, Marioni JC. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Research. 2016; 5(2122). https://doi.org/10.12688/f1000research.9501.2.
Bais AS, Kostka D. scds: computational annotation of doublets in single-cell RNA sequencing data. Bioinformatics. 2020:1150–8. https://doi.org/10.1093/bioinformatics/btz698.
Ilicic T, Kim JK, Kolodziejczyk AA, Bagger FO, McCarthy DJ, Marioni JC, Teichmann SA. Classification of low quality cells from single-cell RNA-seq data. Genome Biol. 2016; 17.1(29):1–15. https://doi.org/10.1186/s13059-016-0888-1.
Kuhn HW. The Hungarian method for the assignment problem. Naval Res Logist Q. 1955; 2(1-2):83–97. https://doi.org/10.1002/nav.3800020109.
L. Lun AT, Bach K, Marioni JC. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 2016; 17(1):75. https://doi.org/10.1186/s13059-016-0947-7.
Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019; 20(1):296. https://doi.org/10.1186/s13059-019-1874-1.
Lin Y, Ghazanfar S, Strbenac D, Wang A, Patrick E, Lin DM, Speed T, Yang JYH, Yang P. Evaluating stably expressed genes in single cells. GigaScience. 2019; 8(9):1–10. https://doi.org/10.1093/gigascience/giz106.
Deeke JM, Gagnon-Bartsch JA. Stably expressed genes in single-cell RNA-sequencing. bioRxiv. 2018:475426. https://doi.org/10.1101/475426.
Bacher R, Chu L-F, Leng N, Gasch AP, Thomson JA, Stewart RM, Newton M, Kendziorski C. SCnorm: robust normalization of single-cell RNA-seq data. Nat Methods. 2017; 14(6):584–6. https://doi.org/10.1038/nmeth.4263. Accessed 02 March 2020.
Lopez R, Regier J, Cole MB, Jordan MI, Yosef N. Deep generative modeling for single-cell transcriptomics. Nat Methods. 2018; 15(12):1053–8. https://doi.org/10.1038/s41592-018-0229-2. Accessed 21 Feb 2019.
Rousseeuw PJ. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math. 1987; 20:53–65. https://doi.org/10.1016/0377-0427(87)90125-7.
Townes FW, Hicks SC, Aryee MJ, Irizarry RA. Feature selection and dimension reduction for single-cell RNA-Seq based on a multinomial model. Genome Biol. 2019; 20(295):1–16. https://doi.org/10.1186/s13059-019-1861-6.
Svensson V, Gayoso A, Yosef N, Pachter L. Interpretable factor models of single-cell RNA-seq via variational autoencoders. Bioinformatics. 2020; 36(11):3418–21. https://doi.org/10.1093/bioinformatics/btaa169.
Johnsson K, Soneson C, Fontes M. Low bias local intrinsic dimension estimation from expected simplex skewness. IEEE Trans Pattern Anal Mach Intell. 2014; 37(1):196–202. https://doi.org/10.1109/TPAMI.2014.2343220.
Chung NC, Storey JD. Statistical significance of variables driving systematic variation in high-dimensional data. Bioinformatics. 2015; 31(4):545–54. https://doi.org/10.1093/bioinformatics/btu674.
Albergante L, Bac J, Zinovyev A. Estimating the effective dimension of large biological datasets using Fisher separability analysis. 2019:1–8. https://doi.org/10.1109/IJCNN.2019.8852450.
Batson J, Royer LA, Webber JT. Molecular Cross-Validation for Single-Cell RNA-seq. bioRxiv. 2019:786269. https://doi.org/10.1101/786269.
Wagner F. Monet: An open-source Python package for analyzing and integrating scRNA-Seq data using PCA-based latent spaces. Preprint. Bioinformatics. 2020. https://doi.org/10.1101/2020.06.08.140673.
Leek JT, Storey JD. Capturing Heterogeneity in Gene Expression Studies by Surrogate Variable Analysis. PLOS Genet. 2007; 3(9):161. https://doi.org/10.1371/journal.pgen.0030161.
Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012; 28(6):882–3. https://doi.org/10.1093/bioinformatics/bts034.
Risso D, Ngai J, Speed TP, Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol. 2014; 32(9):896–902. https://doi.org/10.1038/nbt.2931.
Germain P-L, Vitriolo A, Adamo A, Laise P, Das V, Testa G. RNAontheBENCH: Computational and empirical resources for benchmarking RNAseq quantification and differential expression methods. Nucleic Acids Res. 2016; 44(11):5054–67. https://doi.org/10.1093/nar/gkw448.
Carcamo-Orive I, Hoffman GE, Cundiff P, Beckmann ND, D’Souza SL, Knowles JW, Patel A, Papatsenko D, Abbasi F, Reaven GM, Whalen S, Lee P, Shahbazi M, Henrion MYR, Zhu K, Wang S, Roussos P, Schadt EE, Pandey G, Chang R, Quertermous T, Lemischka I. Analysis of Transcriptional Variability in a Large Human iPSC Library Reveals Genetic and Non-genetic Determinants of Heterogeneity. Cell Stem Cell. 2017; 20(4):518–5329. https://doi.org/10.1016/j.stem.2016.11.005.
The Sequencing Quality Control (SEQC) consortium. A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. Nat Biotechnol. 2014; 32(9):903–14. https://doi.org/10.1038/nbt.2957.
Germain P-L, Testa G. Taming Human Genetic Variability: Transcriptomic Meta-Analysis Guides the Experimental Design and Interpretation of iPSC-Based Disease Modeling. Stem Cell Rep. 2017; 8(6):1784–96. https://doi.org/10.1016/j.stemcr.2017.05.012.
Mereu E, Lafzi A, Moutinho C, Ziegenhain C, MacCarthy DJ, Alvarez A, Batlle E, Sagar Grün D, Lau JK, Boutet S, Sanada C, Ooi A, Jones RC, Kaihara K, Brampton C, Talaga Y, Sasagawa Y, Tanaka K, Hayashi T, Nikaido I, Fischer C, Sauer S, Trefzer T, Conrad C, Adiconis X, Nguyen LT, Regev A, Levin JZ, Janjic A, Wange LE, Bagnoli JW, Parekh S, Enard W, Gut M, Sandberg R, Gut I, Stegle O, Heyn H. Benchmarking Single-Cell RNA Sequencing Protocols for Cell Atlas Projects. bioRxiv. 2019:630087. https://doi.org/10.1101/630087.
Zhang X, Li T, Liu F, Chen Y, Yao J, Li Z, Huang Y, Wang J. Comparative analysis of droplet-based ultra-high-throughput single-cell RNA-Seq systems. Mol Cell. 2019; 73(1):130–42. https://doi.org/10.1016/j.molcel.2018.10.020.
Salomon R, Kaczorowski D, Valdes-Mora F, Nordon RE, Neild A, Farbehi N, Bartonicek N, Gallego-Ortega D. Droplet-based single cell RNAseq tools: a practical guide. Lab Chip. 2019; 19:1706–27. https://doi.org/10.1039/c8lc01239c.
Kim T, Chen IR, Lin Y, Wang AY-Y, Yang JYH, Yang P. Impact of similarity metrics on single-cell RNA-seq data clustering. Brief Bioinforma. 2019; 20(6):2316–26. https://doi.org/10.1093/bib/bby076. Accessed 28 Jan 2020.
Aparicio L, Bordyuh M, Blumberg AJ, Rabadan R. A random matrix theory approach to denoise single-cell data. Patterns. 2020; 1(3):100035. https://doi.org/10.1016/j.patter.2020.100035.
Stuart T, Satija R. Integrative single-cell analysis. Nat Rev Genet. 2019; 20(5):257–72. https://doi.org/10.1038/s41576-019-0093-7.
Saelens W, Cannoodt R, Todorov H, Saeys Y. A comparison of single-cell trajectory inference methods. Nat Biotechnol. 2019; 37(5):547–54. https://doi.org/10.1038/s41587-019-0071-9.
Su S, Tian L, Dong X, Hickey PF, Freytag S, Ritchie ME. CellBench: R/Bioconductor software for comparing single-cell RNA-seq analysis methods. Bioinformatics. 2019. https://doi.org/10.1093/bioinformatics/btz889.
Landau W. The drake R package: a pipeline toolkit for reproducibility and high-performance computing. J Open Source Softw. 2018; 3(21):550. https://doi.org/10.21105/joss.00550.
Frazee AC, Jaffe AE, Langmead B, Leek JT. Polyester: simulating RNA-seq datasets with differential transcript expression. Bioinformatics. 2015; 31(17):2778–84. https://doi.org/10.1093/bioinformatics/btv272.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15(12):550. https://doi.org/10.1186/s13059-014-0550-8.
Lund SP, Nettleton D, McCarthy DJ, Smyth GK. Detecting differential expression in RNA-sequence data using quasi-likelihood with shrunken dispersion estimates. Stat Appl Genet Mol Biol. 2012; 11(5). https://doi.org/10.1515/1544-6115.1826.
Germain P-L, Sonrel A, Robinson MD. Archived code used for publication. figshare. 2020. https://doi.org/10.6084/m9.figshare.12759677.v3. https://figshare.com/articles/software/Archived_code_used_for_publication/12759677/3.
Germain P-L, Sonrel A, Robinson MD. Archived code used for publication. figshare. 2020. https://doi.org/10.6084/m9.figshare.11787210. https://figshare.com/articles/dataset/scRNAseq_benchmark_datasets_with_known_cell_labels/11787210/1.
Funding
This work was supported by the Swiss National Science Foundation (grants 310030_175841, CRSII5_177208) as well as the Chan Zuckerberg Initiative DAF (grant number 2018-182828), an advised fund of Silicon Valley Community Foundation. MDR acknowledges support from the University Research Priority Program Evolution in Action at the University of Zurich.
Author information
Authors and Affiliations
Contributions
PLG developed the pipeComp framework, and PLG and MDR designed the benchmark. PLG and AS prepared the pipelines and method wrappers and performed the analyses. AS performed all denoising/imputation analyses. PLG, AS, and MDR wrote the manuscript. The authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Competing interests
As developers of the scDblFinder package, the authors have an interest in it performing well. The authors declare no further competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1
Supplementary Figures (PDF 16 MB)
Additional file 2
More detailed description of pipeComp and comparison with other tools. (PDF 0.5 MB)
Additional file 3
Review history.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Germain, PL., Sonrel, A. & Robinson, M.D. pipeComp, a general framework for the evaluation of computational pipelines, reveals performant single cell RNA-seq preprocessing tools. Genome Biol 21, 227 (2020). https://doi.org/10.1186/s13059-020-02136-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13059-020-02136-7