Fig. 4
From: Cancer-specific CTCF binding facilitates oncogenic transcriptional dysregulation
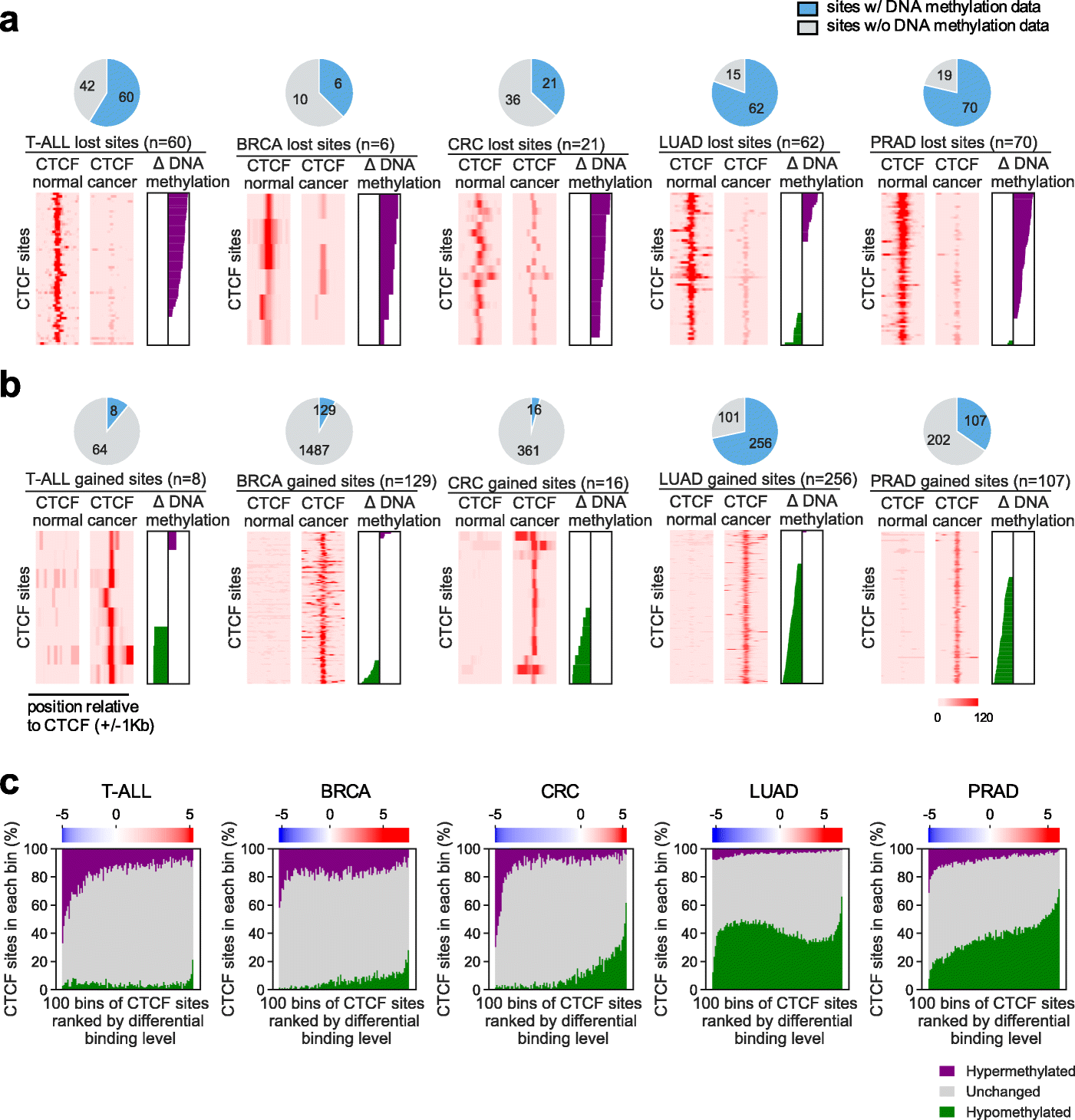
Patterns of differential DNA methylation near cancer-specific lost/gained CTCF sites. a,b ChIP-seq signals and differential DNA methylation levels surrounding specific lost (a) or gained (b) CTCF binding sites in cancer versus normal tissues for each of the 5 cancer types. Pie charts represent numbers of CTCF sites with (light blue) or without (gray) sufficient DNA methylation signals from bisulfite sequencing data. For sites with sufficient DNA methylation signals, heatmaps show CTCF ChIP-seq signals cover 2-kb regions centered at each site. Differential DNA methylation level at a 300 bp region centered at each CTCF binding site was calculated and presented as a waterfall plot. Purple bars represent increased and green bars represent decreased DNA methylation levels (with values in a range from 0 to 100). Rows in corresponding ChIP-seq and DNA methylation plots are ranked identically. c Association between CTCF binding specificity and differential DNA methylation for each cancer type. All CTCF sites with sufficient DNA methylation coverage in each cancer type were ranked based on their differential CTCF binding level in cancer compared to other samples and grouped into 100 equal-count bins. For each bin, the percentages of sites associated with DNA hypomethylation (green), unchanged methylation (gray), and hypermethylation (purple) were stacked in a column. Top color bar represents the median differential binding level of the CTCF sites in a bin, quantified as the T-test statistic
