Fig. 1
From: Mutational signature distribution varies with DNA replication timing and strand asymmetry
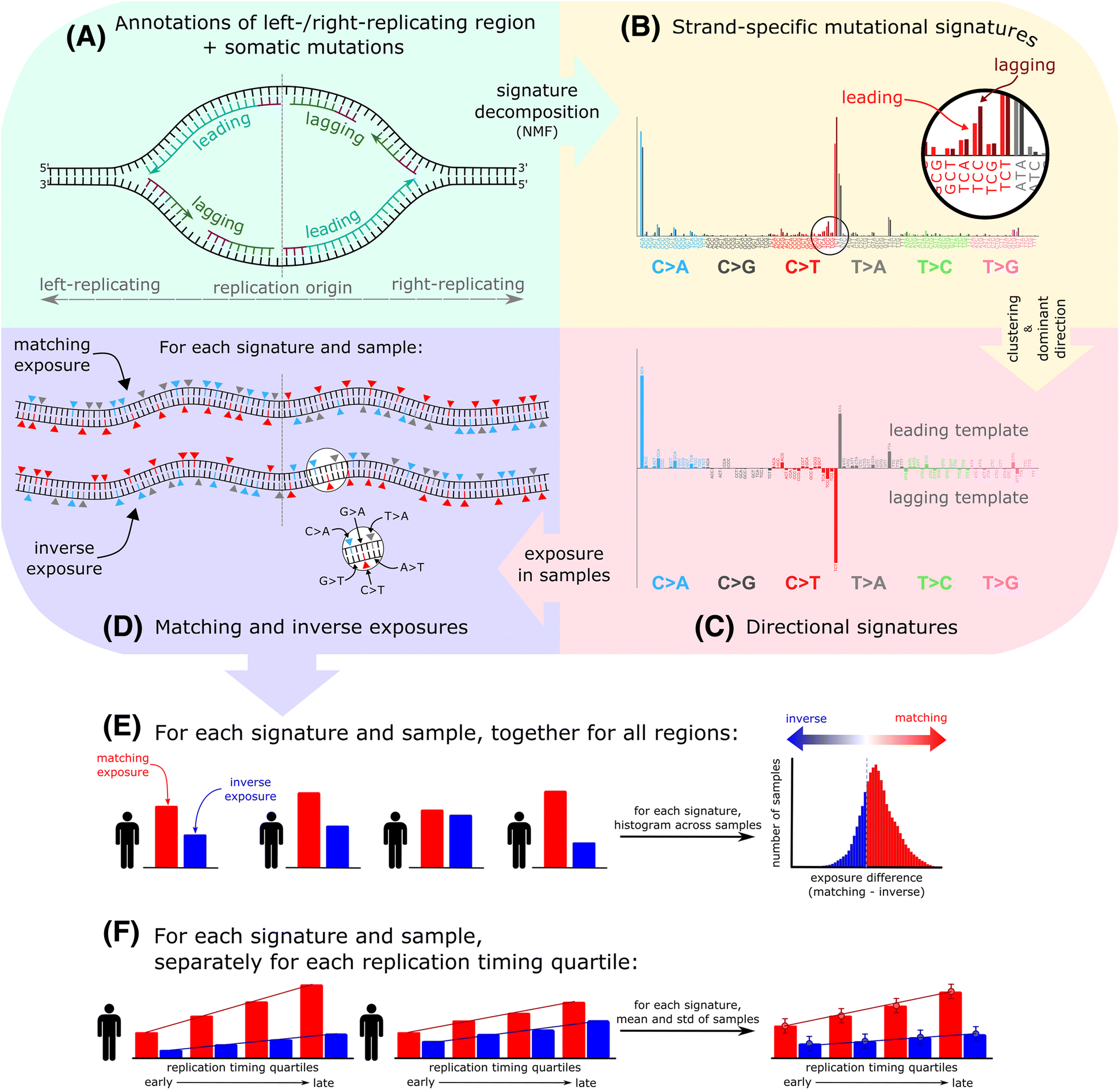
Methods overview. a Mutation frequency on the leading and lagging strands is computed using annotated left/right-replicating regions and somatic single-nucleotide mutations oriented according to the strand of the pyrimidine in the base pair. b Leading and lagging strand-specific mutational signatures are extracted using non-negative matrix factorization. c The signatures are clustered and in each cluster a representative signature is selected (“Methods”). In the cluster representatives, each of the 96 mutation types is annotated according to its dominant direction (upwards-facing bars for leading, downwards-facing bars for lagging template preference). d Exposures to the directional signatures are separately quantified for the leading and lagging strands of each patient. The exposure in the matching orientation reflects the extent to which mutations in pyrimidines on the leading (and lagging) strand can be explained by the leading (and lagging) component of the signature, respectively. Conversely, the exposure in the inverse orientation reflects how mutations in pyrimidines on the leading strand can be explained by the lagging component of the signature (or vice versa) (“Methods”). Top part of d shows an example of a sample with completely matching exposure, given the signature in c, with C > T mutations on the leading template and C > A and T > C mutations on the lagging template, whereas the bottom part of d shows an example of a sample with completely inverse exposure. e Example of matching and inverse exposure quantification in individual patients (for a given signature). Significance of the asymmetry of this signature across the cohort is evaluated based on the distribution of difference between the matching and inverse exposures. The histogram shows an example of a signature with significant matching asymmetry. f Signature exposures are next quantified in bins representing four quartiles of replication timing. The graph on the right shows average and standard deviation values in individual quartiles, representing an example of a signature enriched in the late-replicated regions both in the matching and inverse exposures
