Fig. 1
From: MoDLE: high-performance stochastic modeling of DNA loop extrusion interactions
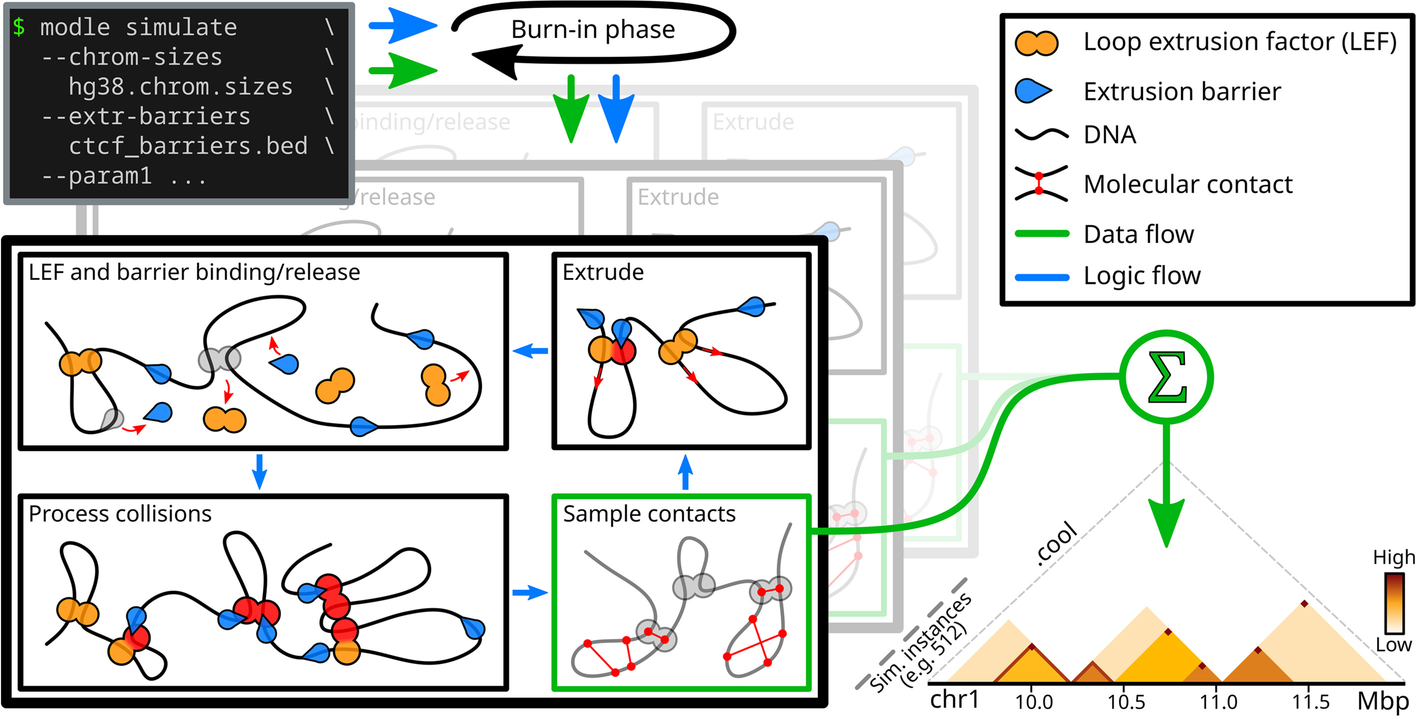
Schematic and simplified overview of MoDLE. Input files specify genome regions to be simulated (e.g., a chrom.sizes file) and their barrier positions (e.g., CTCF binding sites and orientation) in BED format. Optional parameters control the specifics of a simulation. Loop extruding factors (LEFs) bind to, extrude, and release from the regions and interact with modeled barriers according to input parameters. Loop extrusion and intra-TAD contacts of a randomized subset of loops are recorded each epoch and aggregated into an output cooler file containing the final simulated contact frequencies. Simulation halts when a target number of epochs or a target number of loop extrusion contacts have been simulated
