Fig. 1
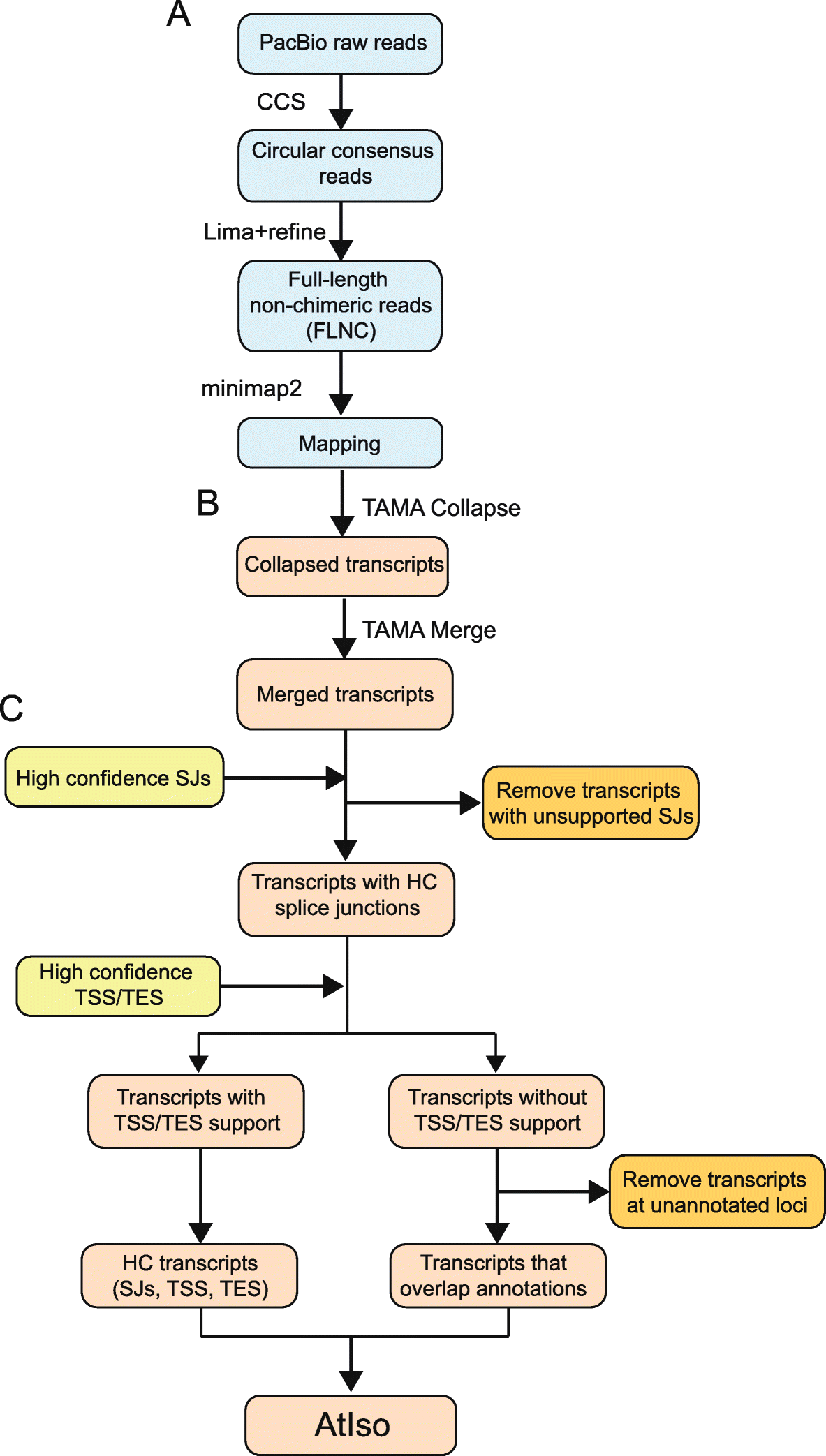
Workflow of analysis of PacBio Iso-sequencing. A Raw reads are analyzed using the PacBio Iso-seq 3 pipeline to generate FLNCs which are mapped to the genome (blue boxes). B Mapped FLNCs are collapsed and merged using TAMA to generate transcripts (pink boxes). C Transcripts are quality controlled using datasets of high-confidence (HC) splice junctions (SJs) and transcript start and end sites (TSS/TES). Transcripts with unsupported splice junctions where reads contain mismatches within ±10 nt of an SJ are removed. Transcripts with both high-confidence TSS and TES (determined by binomial probability for highly expressed genes and by end support with > 2 reads for low expressed genes) are retained as HC transcripts. The remaining transcripts which have partial or no TSS and/or TES support were removed unless they overlapped with annotated gene loci. These transcripts, from genes with low coverage by Iso-seq, were combined with the HC transcripts to form AtIso (Arabidopsis Iso-seq based transcriptome)
