Fig. 1
From: Gramtools enables multiscale variation analysis with genome graphs
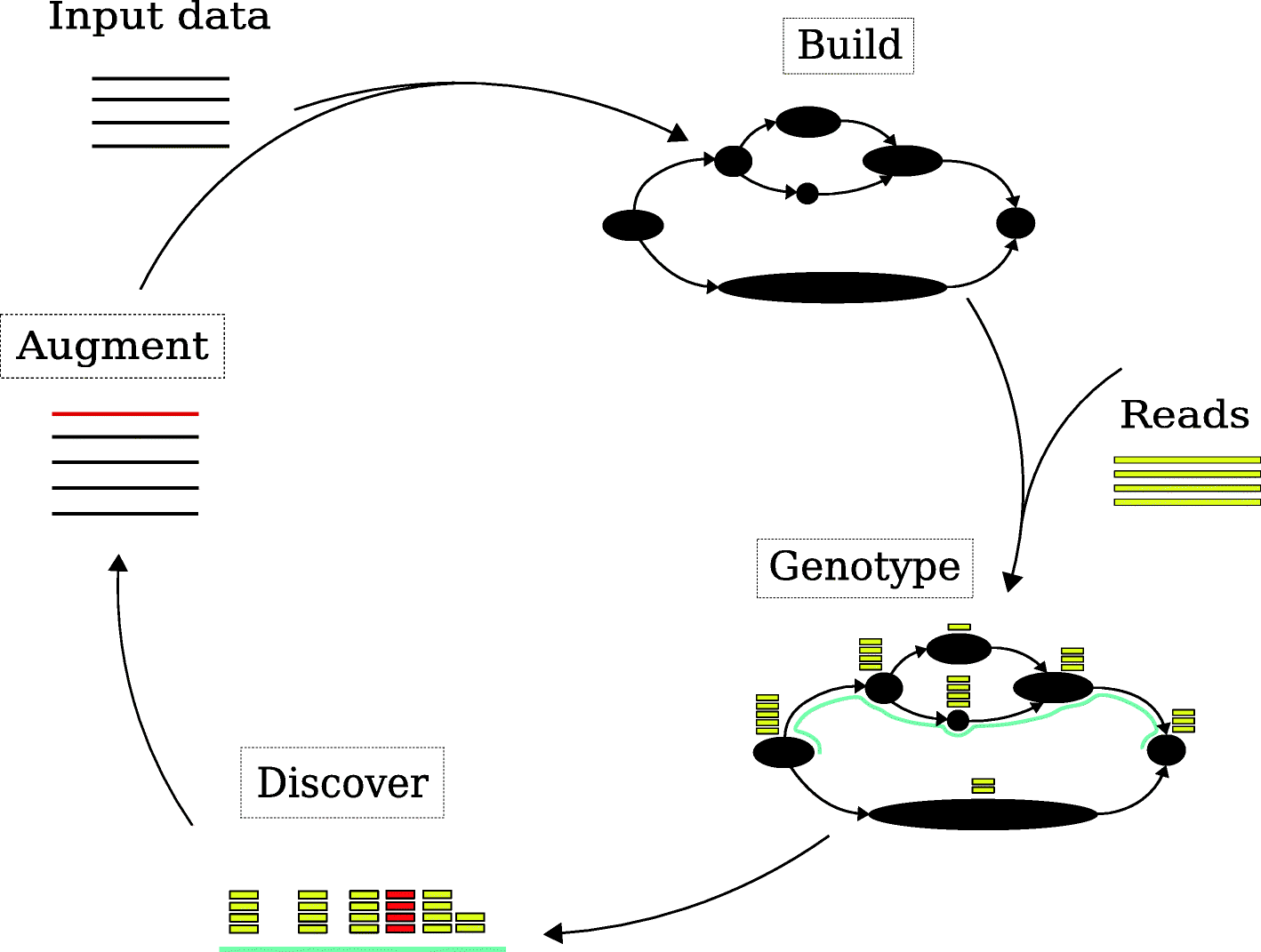
Genome graph workflow implemented in gramtools. Black nodes represent genomic sequence or site entry/exit points. Build consists of producing a genome graph that is directed and acyclic, a requirement for gramtools. Genotyping consists of calling alleles at each variant site and thereby inferring a haploid personalised reference genome for a sample. New variants, shown in red, are discovered using standard reference-based callers run against the personalised reference
