Fig. 6
From: GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes
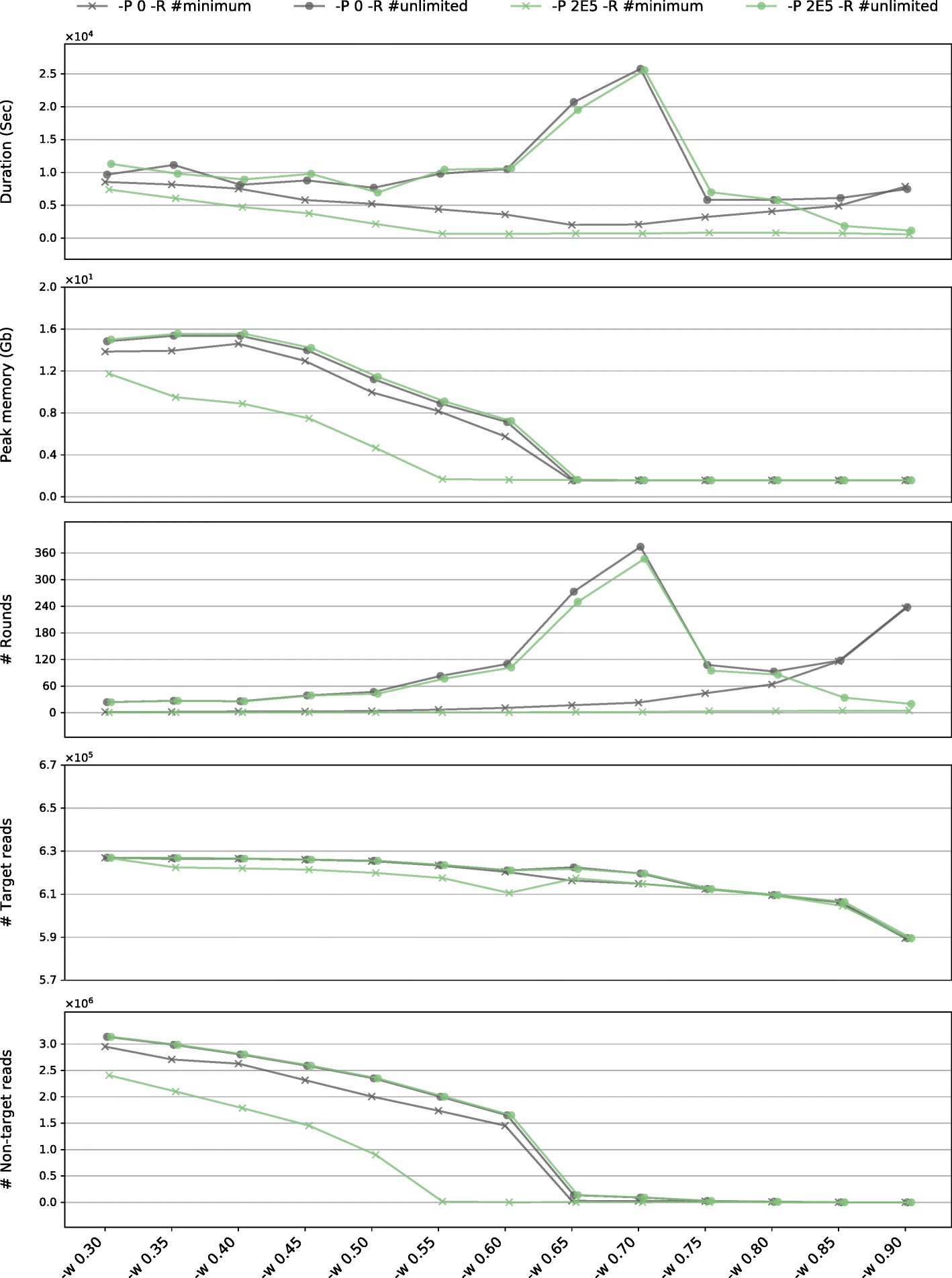
Assessing the plastome assembly performance characteristics of GetOrganelle using online reads of an angiosperm species, Haberlea rhodopensis (SRA: SRR4428742) as the dataset, using the complete plastome of a gymnosperm species, Gnetum parvifolium (GBK: NC_011942.1) as the seed. The assessment was conducted across a range of Word sizes (in the form of WSR, after the flag “-w” along the x axis). The green marks denote using the pre-grouping value 200,000 (after the flag “-P”), while the gray marks denote that the pre-grouping was disabled. The cross marks denote using the minimum number of rounds of extension iterations for achieving a complete plastome or stabilizing the incomplete plastome result, while the solid circle marks denote using unlimited number of rounds (“-R 1000” in practice). From top to bottom, for assembling raw reads into the same complete plastome (or the same incomplete result in all runs of “-w 0.90”), the five subgraphs present the total computational time cost in seconds, the maximum memory cost in gigabytes, the actual number of rounds GetOrganelle took, the number of target plastid reads GetOrganelle recruited, and the number of non-target reads GetOrganelle recruited, respectively
