Fig. 1
From: Genome-wide analysis of DNA replication timing in single cells: Yes! We’re all individuals
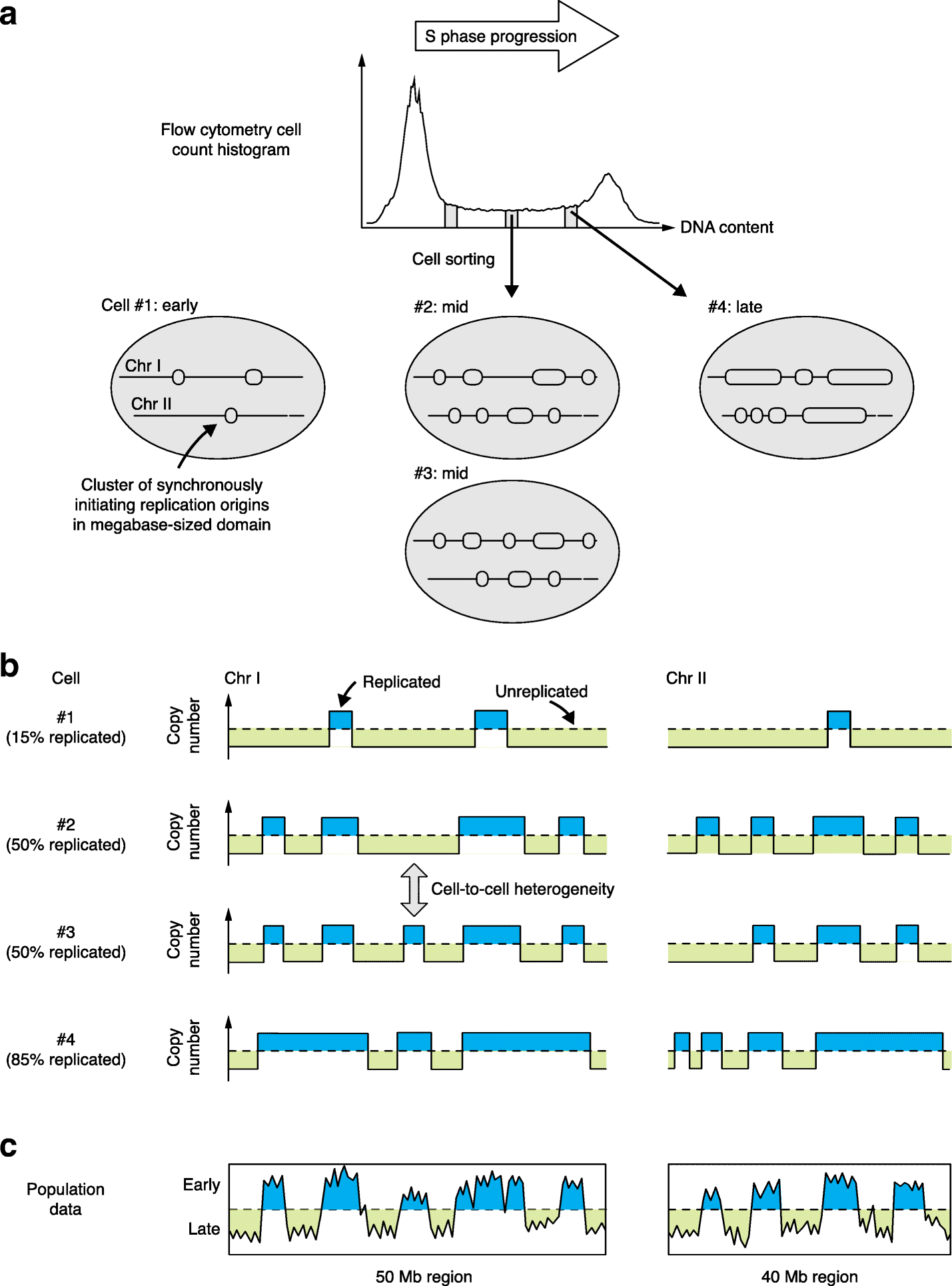
Overview of the procedure used by Takahashi et al. [7] to analyze the replication status of single cells from mouse-derived cell lines. a Individual cells in early, mid or late S-phase were sorted on the basis of their DNA content. The cartoons of cells below illustrate the approximate extent of replication for two chromosome segments in four different cells, cell #1 (early S-phase), cells #2 and #3 (mid S-phase), and cell #4 (late S-phase). Note that these chromosome cartoons illustrate segments of about 50 Mb, with replicated regions of around 1–10 Mb in length that correspond to clusters of activated origins rather than to individual origin sites. b Principle of replication status analysis in single cells. Sequences are assigned as replicated (Copy number 2; blue fill) or not (Copy number 1; green fill) on the basis of their representation in the high-throughput sequencing analysis of each single cell, as illustrated in the stylized plots shown for each cell. The sequence data also allow assignment of the percentage of the genome that is replicated in each cell (shown on the left). c Plots illustrate the replication timing curves that would be obtained for the same chromosome segments using traditional genome-wide replication timing analysis in a large cell population (such as population Repli-seq as described in [7]); the results are expected to resemble mid-S-phase copy number plots (i.e. cells #2 and #3) most closely
