Fig. 2
From: Comparative analysis of sequencing technologies for single-cell transcriptomics
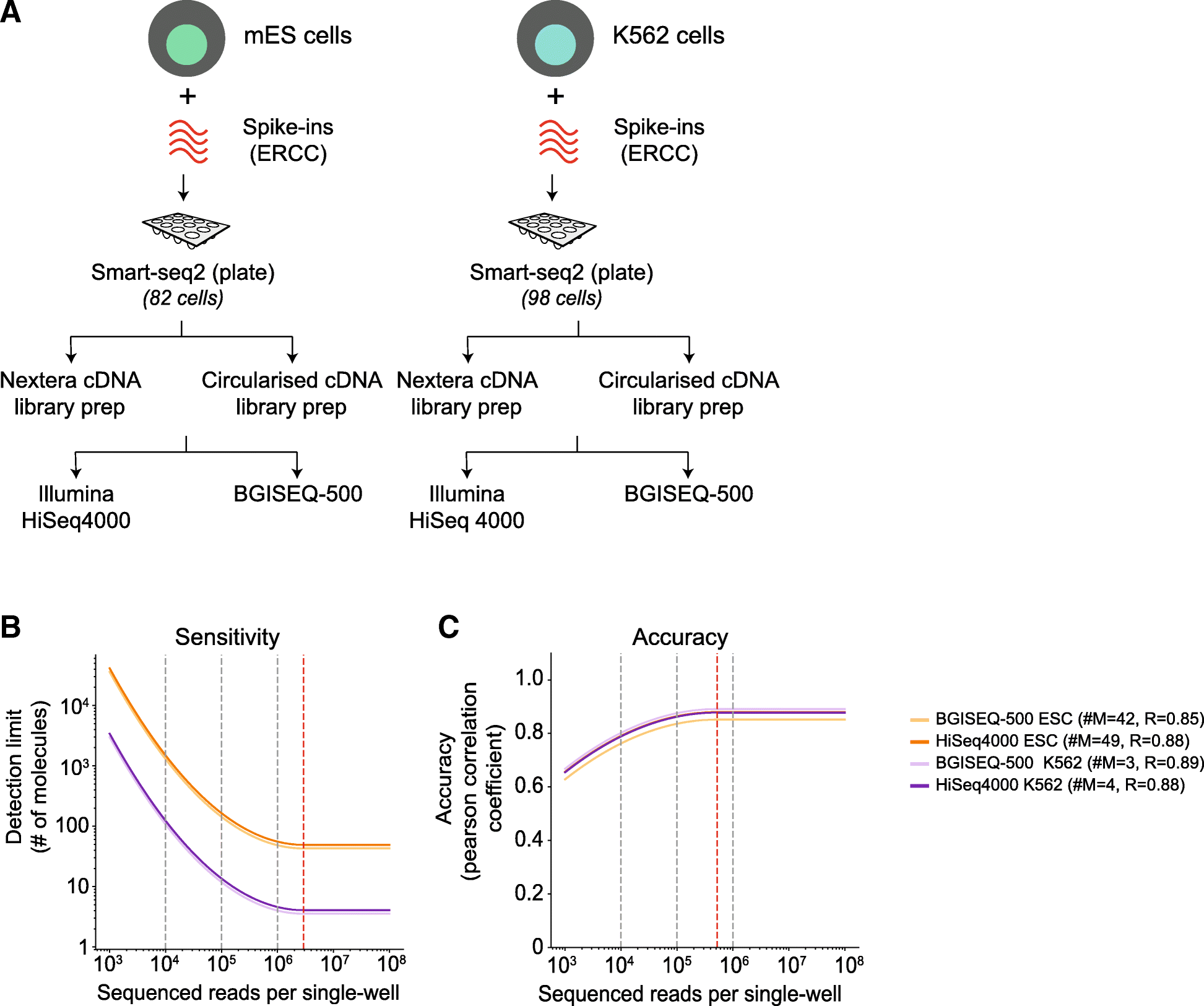
a Schematic overview of the mESC and K562 scRNA-seq experiment using plate-based Smart-seq2 protocol and sequencing. 82 mESCs and 98 K562s are profiled using plate-based Smart-seq2 protocols, followed by matched single- and paired-end library preparation for both Illumina and BGISEQ-500 platform resulting in 721 matched libraries. b The sensitivity of mESCs and K562s cells downsampled across two orders of magnitude. The sensitivity is critically dependent on sequencing depth and deeply sequenced K562 cells appear to be more sensitive than mESCs. The sensitivity, however, between the sequencing platforms is highly similar. c The accuracy of mESCs and K562s downsampled across two orders of magnitude. The accuracy has little dependence on sequencing depth, and both mESCs and K562 have similar accuracies, across both sequencing platform. The grey dotted lines indicate downsampling at different read depths per cell, while red line indicates saturation per cell
