Fig. 9
From: Skmer: assembly-free and alignment-free sample identification using genome skims
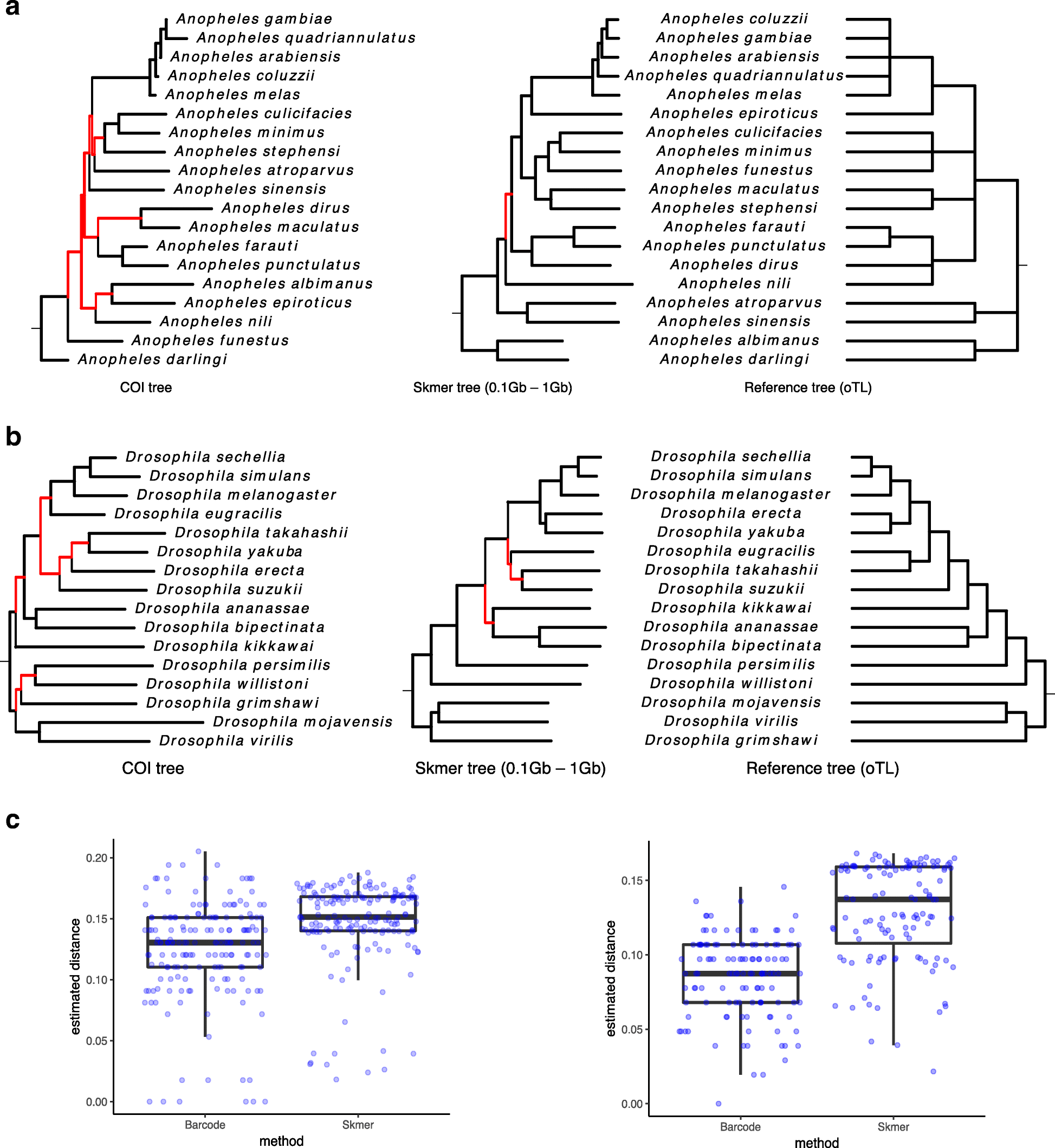
Comparing distances and phylogenetic trees from COI barcodes and simulated genome skims. Shown in red are wrong internal branches corresponding to the bipartitions that are not found in the reference tree. Genome-skim size is randomly chosen among 0.1 Gb, 0.5 Gb, and 1 Gb. aAnopheles trees. bDrosophila trees. c Distribution of distances for Anopheles (left) and Drosophila (right) genomes
