Fig. 1
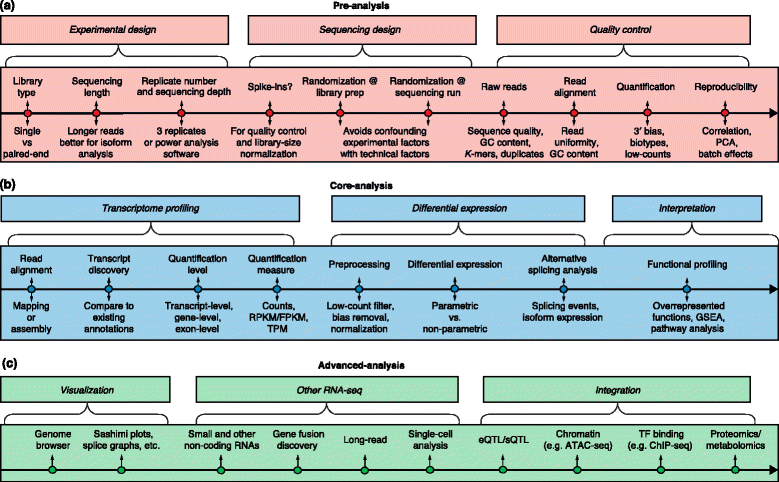
A generic roadmap for RNA-seq computational analyses. The major analysis steps are listed above the lines for pre-analysis, core analysis and advanced analysis. The key analysis issues for each step that are listed below the lines are discussed in the text. a Preprocessing includes experimental design, sequencing design, and quality control steps. b Core analyses include transcriptome profiling, differential gene expression, and functional profiling. c Advanced analysis includes visualization, other RNA-seq technologies, and data integration. Abbreviations: ChIP-seq Chromatin immunoprecipitation sequencing, eQTL Expression quantitative loci, FPKM Fragments per kilobase of exon model per million mapped reads, GSEA Gene set enrichment analysis, PCA Principal component analysis, RPKM Reads per kilobase of exon model per million reads, sQTL Splicing quantitative trait loci, TF Transcription factor, TPM Transcripts per million
