Figure 1
From: Pervasive and dynamic protein binding sites of the mRNA transcriptome in Saccharomyces cerevisiae
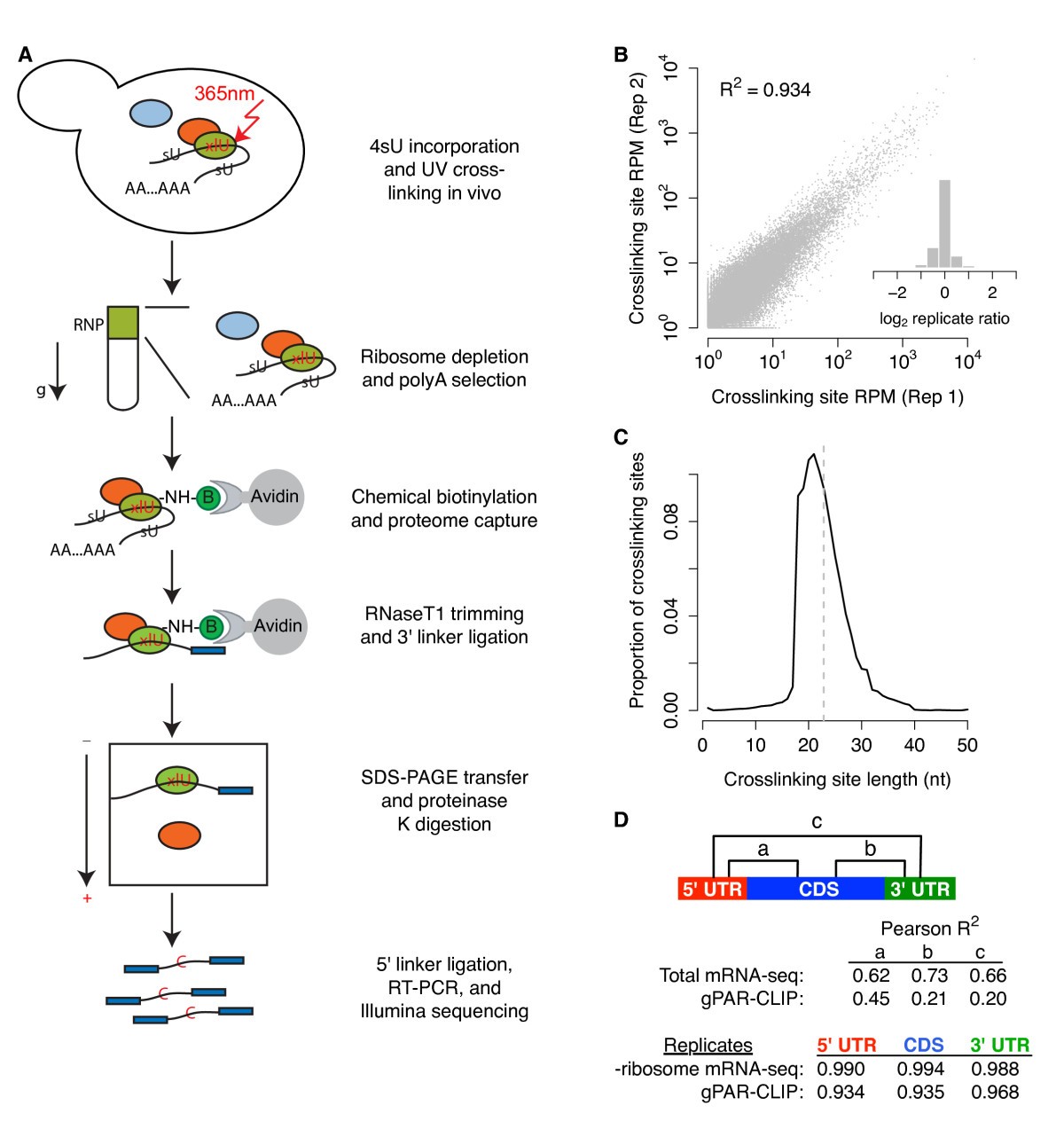
gPAR-CLIP identifies transcriptome-wide RBP crosslinking sites. (a) Schematic of the gPAR-CLIP protocol. (b) Reproducibility of crosslinking sites generated from replicate gPAR-CLIP libraries prepared from yeast grown in synthetic defined media (abbreviated as WT gPAR-CLIP hereafter). Pearson correlation coefficient is indicated. Inset: distribution of log2 crosslinking site RPM ratios between replicates. Replicate error σ = 1.3-fold. (c) Length distribution of crosslinking sites in WT gPAR-CLIP libraries. Dotted line: average crosslinking site length of 23 nucleotides. (d) Pearson correlation coefficients of total mRNA-seq and gPAR-CLIP read coverage between 5' UTR, CDS, and 3' UTR regions as well as correlation coefficients of ribosome depleted (-ribosome) mRNA-seq and gPAR-CLIP read coverage between replicate WT libraries.
