Figure 1
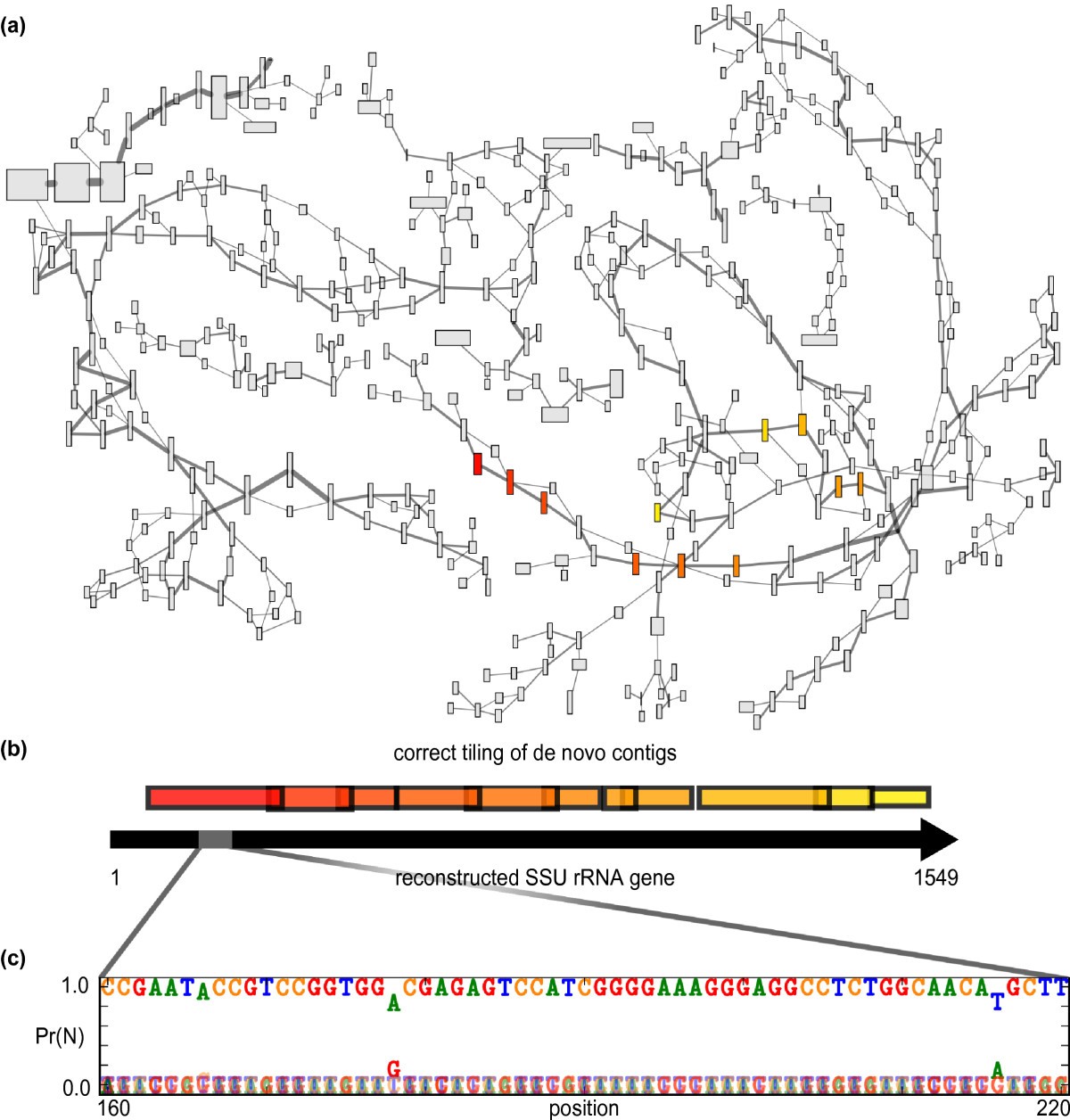
De novo assembly of SSU rRNA genes versus reconstruction of full-length gene sequences. (a) A section of the de Bruijn graph created by the short read assembler Velvet [29] for the natural microbial community. Each contig in the graph is represented by a rectangle whose width is proportional to contig length and whose height is proportional to contig k-mer coverage depth. Edge width reflects the multiplicity of overlapping k-mers shared by contigs. All contigs with BLAST matches to SSU genes recovered by EMIRGE were selected, and those contigs and additional contigs within three edges are shown. Contigs with BLAST matches to the SSU sequence from Leptospirillum ferrodiazotrophum [54] are shown in color. (b) The correct tiling of highlighted contigs from (a) is shown schematically with the EMIRGE-reconstructed SSU rRNA gene. (c) A selected region of the L. ferrodiazotrophum SSU gene shows the individual base probabilities at algorithm termination for each position in the reconstructed SSU gene. While most bases are highly confident, some positions show evidence for strain variants present in the population.
