Fig. 1
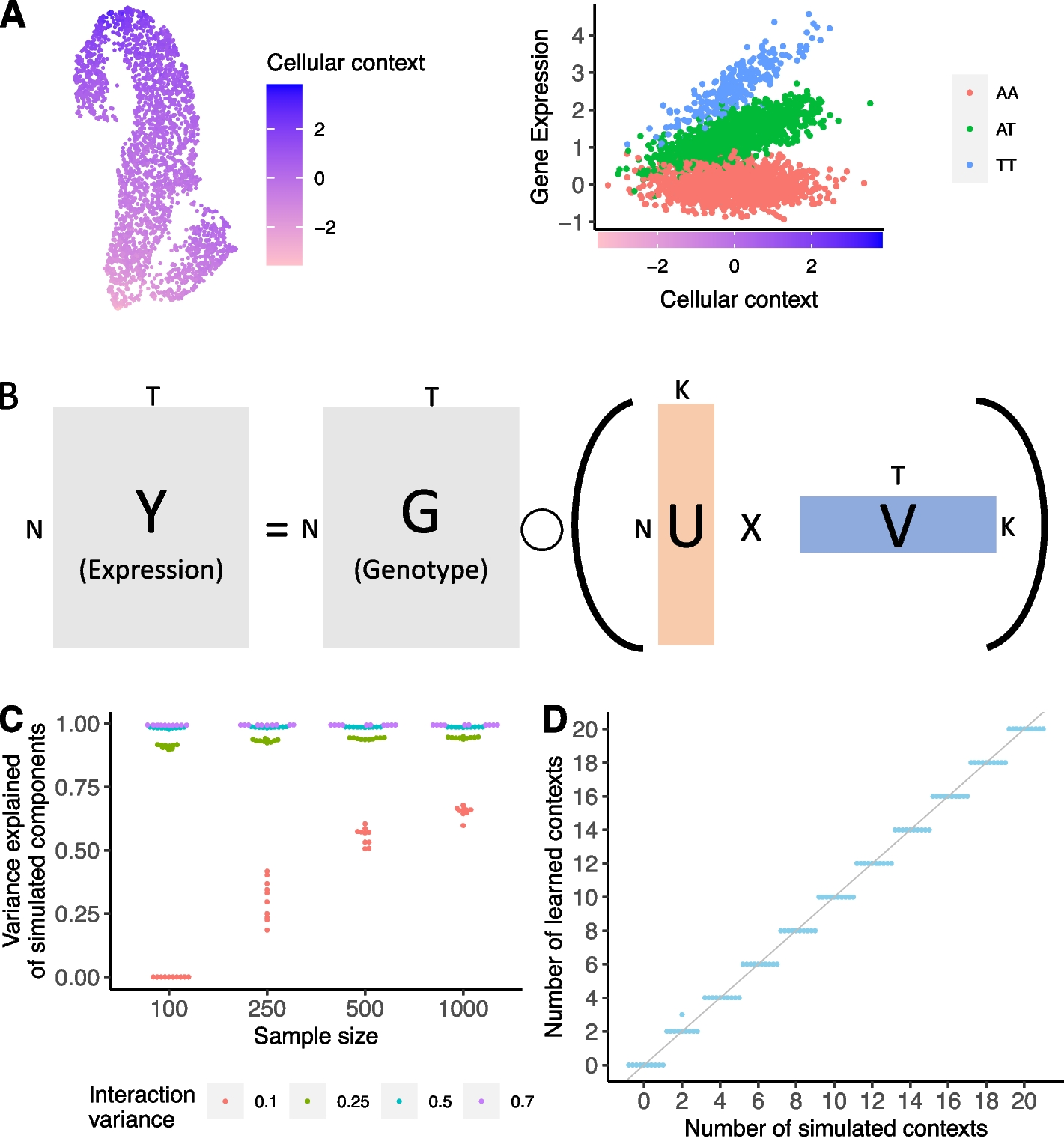
SURGE model overview and simulation. A Schematic example of an interaction eQTL where the eQTL effect size (right) changes as a function of cellular context (depicted in UMAP embedding, left). B SURGE is a novel probabilistic model that uses matrix factorization to jointly learn a continuous representation of the cellular contexts defining each measurement (U) and the corresponding eQTL effect sizes specific to each learned context (V) based on observed expression (Y) and genotype (G) data. SURGE additional accounts for the effects of known covariates and sample repeat structure on gene expression (not shown in figure; see the “Methods” section). Assume there are N samples, T genome-wide independent variant-gene pairs, and K latent contexts. C Based on simulated data, we evaluated SURGE’s ability to reconstruct simulated latent contexts as measured by the average variance explained of the simulated latent contexts by the learned latent contexts (y-axis). We simulate 5 latent contexts and vary the sample size (x-axis) and the strength (variance; see the “Methods” section) of the interaction terms (colors). We fix the fraction of tests that are context-specific eQTLs for each context to .3 (see the “Methods” section). For each parameter setting, we run 10 independent simulations. Each dot is an independent simulation. D Based on simulated data, we evaluate SURGE’s ability to identify the number of simulated latent contexts across 10 independent simulations. The sample size was fixed to 250, the strength (variance) of the simulated interaction terms was fixed to .25, and the fraction of tests that are context-specific eQTLs for a particular context (see the “Methods” section) was fixed to .3. For each parameter setting, we run 10 independent simulations. Each dot is an independent simulation
